ISSN: 0973-7510
E-ISSN: 2581-690X
The advent of technologies has made allogenic transplantation a potential curative therapy for end-stage renal diseases, but the episodes of rejection still remain as one of the challenges in the post-transplant scenario. In the recent years, several human and animal studies have elucidated that gut microbial dysbiosis is closely linked with allogenic transplantation and post-transplant complications. But most of the studies focused on the use of high through-put sequencing technologies to analyze gut microbiota despite of its high cost, analysis and time constraints. Hence, in this work we aimed to study the impact of the two dominant gut phyla Firmicutes and Bacteroidetes on 38 renal transplant recipients, before and after transplantation and to find its association with allograft rejection. Significant changes (p<0.01) were observed in the relative abundances of the phyla Firmicutes and Bacteroidetes at pre- and post-transplant period. We have also found that the recipients who had an increase in Firmicutes/Bacteroidetes (F/B) ratio before transplant were highly prone to rejection in the first-year post-transplant. The Receiver Operating Characteristic (ROC) curve analysis has shown that the ratio of F/B were able to discriminate between rejection and non-rejection cases with an Area under the ROC Curve (AUC) of 0.91. Additionally, we observed that the ratio of F/B have reduced during the time of rejection postulating that gut microbial dysbiosis has more association with rejection. Thus, the assessment of F/B ratio using qPCR would be of a more practical approach for diagnosis and monitoring of graft function in a cost-effective and timely manner.
Gut Microbiota, Firmicutes, Bacteroidetes, Renal Transplantation, Renal Allograft Rejection, qPCR
Allogenic renal transplantation is a potential therapeutic tool for end-stage renal diseases but post-transplant complications such as rejection, infection, graft versus host disease (GVHD) and relapse remain as a challenge in maintaining long-term graft function. Various evidences have stated that post-transplant complications are closely related to immune system,1 but a dynamic interplay between the host immune system and gut microbiota,2,3 further elaborates the association of gut microbiota with allogenic transplantation.4,5
The human gut microbiota imparts specific function in the host nutrient metabolism, xenobiotic and drug metabolism, maintenance of structural integrity of the gut mucosal barrier, immunomodulation and protection against pathogens.6 The normal human gut microbiota comprises of two dominant bacterial phyla Firmicutes and Bacteroidetes that constitute over 90% of the known phylogenetic categories7 followed by other subdominant phyla including Proteobacteria, Actinobacteria, Fusobacteria, Tenericutes and Verrucomicrobia. Phylum Firmicutes contains Gram-positive bacteria with rigid or semi-rigid cells walls that are largely from the genera Bacillus, Clostridium, Enterococcus, Lactobacillus, and Ruminococcus8 whereas phylum Bacteroidetes includes approximately 7000 different species of Gram-negative bacteria that are predominantly from the genera Bacteroides, Alistipes, Parabacteroides, and Prevotella.9
The disturbances in the microbial community characterized by a loss or gain of microbes that either promote health or diseases is termed as dysbiosis.10 Gut microbial dysbiosis drives a series of abnormalities like accumulation of uremic toxins, systemic inflammation and infection leading to kidney diseases. In turn these renal pathological conditions can affect the microbial community leading to dysbiosis.11-15 Thus, a balance in the gut microbial diversity is essential to maintain an immunological balance. Among the microbial communities, the ratio of the predominant rulers, Firmicutes and Bacteroidetes tend to have an important influence in the maintenance of normal intestinal homeostasis and hence changes in this ratio leads to various pathologies.16 Thus, our main focus of this study is to examine whether there are any changes in the ratio of Firmicutes/Bacteroidetes following renal transplantation and whether the changes in the ratio leading to dysbiosis is connected to post-transplant allograft rejection.
Despite of the popularity of Next Generation Sequencing (NGS) to analyze the changes in gut microbiota, we have used quantitative real-time Polymerase Chain Reaction (qPCR) to estimate the relative abundance of Firmicutes, Bacteroidetes, Gamma Proteobacteria and Epsilon Proteobacteria with an intention to pave way for its translational utility, as qPCR is more rapid, sensitive and a cost-effective method whereas NGS is costly, requires trained persons to analyze data and time consuming restricting its translational purpose.
Study design
It is a longitudinal cohort study comprising of 38 renal transplant recipients who have given written informed consent to participate in the study. The research work has got ethical clearance from Institute’s Human Ethical committee (JIP/IEC/2017/0115 dated 25th May, 2017) and the work has been conducted during the period 2018-2020, in accordance with the principles set forth in Helsinki declaration.
Study cohort
Our 38 renal transplant recipients had a live related ABO and human leucocyte antigen (HLA) matched donor for transplantation. The recipients were under induction therapy with recombinant anti-thymocyte globin (R-ATG) 1.5 mg/kg body weight and 20 µg of Basiliximab for two days prior to transplantation. The post-transplant immune suppression regimen consists of tacrolimus 0.1 mg/kg body weight, 1 g of Mycophenolate mofetil and 20 mg prednisolone. Appropriate changes were made in the maintenance regimen based on the patients’ clinical findings and laboratory work up during the follow-up period. All of the study participants were clinically followed up to two years post-transplant and recipients who had graft dysfunction during this period underwent renal biopsy. The histopathological assessment of renal biopsy was done using Banff 2018 criteria17 based on which we categorized our cohort as those with rejection and those without rejection.
Sampling
The study participants were instructed to collect fresh faecal sample in sterile sealed container and submit the specimen as early as possible (~ within 4 hours) of collection.18,19 The specimens were transported to the laboratory at room temperature18,20 as all of our participants were in proximity. Faecal specimen collection was made at three time points viz, pre-transplant (before the initiation of induction therapy), one month and three months post-transplant.
Nuclei acid extraction
The faecal specimen was immediately subjected for DNA isolation with QIAamp DNA stool mini kit (catalogue no: 51604). The isolated DNA was checked for purity using Qubit 3.0 fluorometer and assay kits (Invitrogen- Q32852, Q32851, Q33211).
Quantification using real-time PCR
Equal concentrations of DNA (1 ng/µl) were used for the quantification of Phylum Bacteroidetes, Phylum Firmicutes, Class Gamma Proteobacteria and Class Epsilon Proteobacteria. qPCR amplification and detection were performed using QuantStudio3 thermal cycler (Applied Biosystems). Each PCR reaction consists of 10µl TaqMan Fast advanced PCR mix (Applied Biosystems – 4444556), 5 µl of DNA, 1 µl of specific TaqMan probe (Customized with FAM dye by Applied Biosystems – 4331348), 1 µl of 16S rRNA (Customized with VIC dye by Applied Biosystems – 4331348) TaqMan probe and 3 µl of Nuclease free water (Qiagen – 129115) making up to a final volume of 20 µl/well. The assay Id’s for the customized TaqMan probe are listed (Table1). The amplification program consists of Uracil-N-glycosylase (UNG) incubation at 50°C for 2 minutes, polymerase activation at 95°C for 20 seconds and 40 cycles of denaturation and annealing at 95°C for 1 second and 60°C for 20 seconds, respectively. The relative amount of Bacteroidetes, Firmicutes, Gamma Proteobacteria and Epsilon Proteobacteria in each sample were normalized to the 16S rRNA universal primer.
Table (1):
Assay Id’s of the TaqMan probes for Phylum Bacteroidetes, Firmicutes and Class Gamma Proteobacteria, Class Epsilon Proteobacteria and 16SrRNA (Universal Primer) customized by Applied Biosystems.
Taxonomy |
Assay ID |
|---|---|
Phylum Bacteroidetes |
APFVNUR |
Phylum Firmicutes |
APGZHEN |
Class Gamma Proteobacteria |
APU666X |
Class Epsilon Proteobacteria |
APWCZRV |
16S rRNA |
AP7DUTJ |
Statistical analysis
Shapiro-Wilk normality test was used for checking the distribution of data. The variables used in this study did not follow a normal distribution and hence non-parametric tools were used for the analysis. Continuous variables have been expressed as either mean ± standard deviation or as median and interquartile range (Q1 to Q3). Friedman’s test was used to find the changes of relative abundances of the microbiota between pre- and post-transplant period. Post-hoc analysis with Wilcoxon signed rank test was conducted with a Bonferroni correction to estimate the time where the actual significant changes of relative abundance occurred. Mann-Whitney U test was used to compare between rejection and non-rejection groups. Logistic regression analysis was done to estimate the association of F/B ratio with rejection. ROC curve was plotted and the AUC along with 95% Confidence Interval (CI) was estimated to find the predictive performance of the F/B ratio in differentiating rejection from non-rejection. SPSS version 19 was used for all statistical analysis. Significant threshold was set at p<0.05.
Rejection outcome
Thirteen of our 38 renal transplant recipients had a biopsy proven rejection within a time span of 15 days to 12 months post-transplant. Out of these 13 with rejection, 11 of them had only Antibody mediated rejection (ABMR) and the other two recipients with ABMR also had T cell and borderline rejection. At the end of follow-up, we have found that eight of the 13 recipients who had rejection episode had a poor graft survival with an eGFR>60mL/ min/1.73m2). The baseline characteristics of our transplant cohort is listed in Table 2.
Table (2):
Characteristics of the renal transplant cohort represented as mean ± standard deviation or median (Q1, Q3).
Characteristics |
Variables |
|---|---|
Recipients’ age (years) |
36.29 ± 9.14 |
Donor’s age (years) |
41.88 ± 11.82 |
Recipients’ gender |
94.7% male and 5.3% female |
Donor’s gender |
24% male and 66% female |
Recipients’ cold ischemia time (minutes) |
88 (70,120) |
Recipients’ warm ischemia time (minutes) |
5.92 ± 2.07 |
Recipients’ BMI (kg/m2) |
20.09 ± 4.73 |
Recipients’ blood pressure Systolic/diastolic (mmHg) |
134 (131,150)/90 (80,90) |
Pre- and post-transplant microbial changes
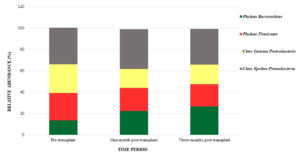
The relative abundance of the phylum Firmicutes, phylum Bacteroidetes, class Gamma Proteobacteria and class Epsilon bacteria were measured before transplant and one and three months after transplant (Figure 1). The pre- and post-transplant changes in the relative abundance of the microbiota are tabulated (Table 3). It was observed that the phylum Firmicutes were more and Bacteroidetes were less before transplant and hence the F/B ratio was more pre-transplant. We have also found that the relative abundance of Gamma Proteobacteria was more in the pre-transplant period than the post-transplant period, whereas the relative abundance of Epsilon Proteobacteria was less in pre-transplant than the post-transplant period and these findings were statistically significant. The post-hoc analysis with Wilcox signed rank test revealed that there were significant changes (p<0.01) in the relative abundance of phylum Bacteroidetes and Firmicutes between pre and one-month, pre and three-months and one and three-months whereas in class Gamma Proteobacteria the changes were not significant between one and three-months and in Class Epsilon Proteobacteria the changes between pre and three-months were not found to be significant.
Table (3):
The changes in the relative abundance at each time point represented as median values (Q1, Q3).
Taxonomy |
Pre-transplant |
One-month post-transplant |
Three-months post-transplant |
p-value |
|---|---|---|---|---|
Phylum Bacteroidetes |
13.77 (9.98, 15.49) |
22.32 (18.29, 25.41) |
26.82 (20.07, 30.31) |
<0.01 |
Phylum Firmicutes |
25.50 (21.50, 28.60) |
21.63 (19.50, 24.59) |
20.45 (18.03, 23.01) |
<0.01 |
Firmicutes / Bacteroidetes ratio |
1.96 (1.50, 2.68) |
0.95 (0.86, 1.23) |
0.80 (0.62, 1.14) |
<0.01 |
Class Gamma Proteobacteria |
26.82 (23.02, 28.49) |
17.66 (15.46, 21.99) |
18.57 (16.05, 24.26) |
<0.01 |
Class Epsilon Proteobacteria |
34.22 (32.39, 37.11) |
37.35 (35.78, 38.47) |
33.37 (31.77, 36.67) |
<0.01 |
Figure 1. Changes in the relative abundances of Phyla Firmicutes, Bacteroidetes, Class Gamma and Class Epsilon Proteobacteria across time period.
Association of Pre-transplant F/B ratio with allograft rejection
Recipients who had a higher relative abundance of Firmicutes, resulting in a higher F/B ratio before their transplantation experienced rejection within one-year post-transplant than those who had less F/B ratio before their transplant (Table 4). We further constructed a logistic regression model to assess the ability of F/B ratio to predict rejection. Preliminary analyses were performed to ensure there was no violation of assumptions. The model was able to classify 78.9% of cases and there was a high association between F/B ratio and rejection outcome with an odds ratio of 9.00 (95% CI, 1.58-51.20) and significance of p<0.05 (Table 5).
Table (4):
Pre-transplant relative abundances of the taxonomic groups between rejection and non-rejection groups represented as median values (Q1, Q3).
Taxonomy |
Rejection |
Non-Rejection |
p-value |
|---|---|---|---|
Phylum Bacteroidetes |
8.55 (5.31, 13.81) |
14.66 (10.82, 16.78) |
<0.01 |
Phylum Firmicutes |
32.28 (28.54, 33.56) |
23.82 (20.88, 25.50) |
<0.01 |
Firmicutes / Bacteroidetes ratio |
3.68 (2.08, 6.23) |
1.65 (1.29, 2.01) |
<0.01 |
Class Gamma Proteobacteria |
26.15 (21.17, 28.25) |
26.99 (23.22, 29.23) |
0.22 |
Class Epsilon Proteobacteria |
34.36 (31.76, 36.90) |
34.07 (32.15, 38.13) |
0.62 |
Table (5):
Logistic regression analysis to establish the relation between the pre-transplant relative abundance of taxa and rejection outcome.
Variable |
β0 |
β1 |
Wald Statistic (β1) |
OR (95% CI) |
p-value |
|---|---|---|---|---|---|
Phylum Bacteroidetes |
3.61 |
-0.35 |
7.83 |
0.70 (0.55, 0.90) |
0.01 |
Phylum Firmicutes |
-38.74 |
1.4 |
5.1 |
4.05 (1.20, 13.61) |
0.02 |
Firmicutes / Bacteroidetes ratio |
-5.67 |
2.2 |
6.13 |
9.00 (1.58, 51.20) |
0.01 |
Class Gamma Proteobacteria |
3 |
-0.14 |
1.99 |
0.87 (0.71, 1.06) |
0.16 |
Class Epsilon Proteobacteria |
1.68 |
-0.07 |
0.5 |
0.94 (0.78, 1.18) |
0.48 |
Diagnostic ability of Pre-transplant F/B ratio
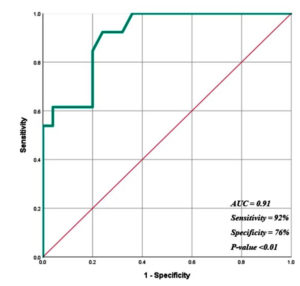
ROC analysis was done to check the performance of F/B ratio in classifying rejection and non-rejection cases. We have found that the F/B ratio had a higher level of diagnostic capability to predict rejection and non-rejection outcomes with an AUC of 0.91 (95%CI, 0.82, 1.0), sensitivity of 92% and specificity of 76% with a significance p<0.01 (Figure 2).
Temporal association of F/B ratio with allograft rejection
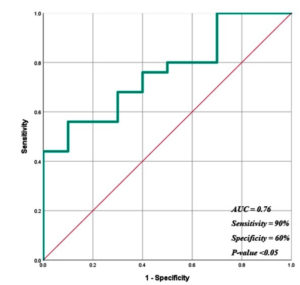
At the time of rejection, the relative abundance of Firmicutes were less and Bacteroidetes were more thus leading to a decrease in the F/B ratio in the rejection group in comparison to non-rejection group (Table 6) (p<0.05). The logistic regression model revealed that there was a significant association (p<0.05) between F/B ratio and rejection outcome with a classification accuracy of 74.3%, AUC of 0.76 (95%CI, 0.61-0.93) and a sensitivity and specificity of 90% and 60%, respectively (Figure 3).
Table (6):
Differences in the relative abundance between rejection and non-rejection groups at the time of rejection.
Taxonomy |
Rejection |
Non-Rejection |
p-value |
|---|---|---|---|
Phylum Bacteroidetes |
25.24 (23.62, 28.23) |
19.14 (17.64, 25.91) |
0.03 |
Phylum Firmicutes |
20.75 (20.20, 20.27) |
21.41 (17.57, 23.94) |
0.91 |
Firmicutes / Bacteroidetes ratio |
1.94 (1.42, 2.62) |
2.70 (1.97, 3.46) |
0.02 |
Class Gamma Proteobacteria |
15.96 (15.04, 18.16) |
18.96 (16.18, 23.22) |
0.01 |
Class Epsilon Proteobacteria |
35.13 (32.03, 36.86) |
35.18 (32.86, 36.38) |
0.93 |
The ratio of F/B has been suggested as an index of health of gut microbiome although they have high inter-individual variations and dramatic dynamics.21 Hence, in our study we intend to study the alterations in the relative abundances of Phyla Bacteroidetes, Firmicutes, Class Gamma Proteobacteria and Epsilon Proteobacteria following renal transplantation and investigated the use of F/B ratio to characterize allograft rejection.
The faecal specimens collected from our 38 renal transplant recipients at three time periods (pre-transplant, one- and three-months post-transplant) revealed that the relative abundance of phylum Firmicutes were more and phylum Bacteroidetes were less at pre-transplant leading to an increase in the ratio of F/B. This finding is similar to the findings reported by Lee et al., wherein they found a higher abundance of Firmicutes to Bacteroidetes in the five faecal specimens of the renal transplant recipients before their transplantation.22 The human microbiome consortium has characterized that a healthy individual has more Bacteroidetes than Firmicutes,23 hence an increase in the phylum Firmicutes in our report states that there is a dysbiosis in the F/B ratio even before transplant when the patients are in End-Stage Renal Disease (ESRD) stage. Our result was further supported by Vaziri et al., who analysed the faecal specimen from 24 ESRD patients and 12 healthy volunteers and found an increase in the number of Firmicutes in ESRD group than the control group.24
In the year 2014, Fricke et al.,25 longitudinally investigated the changes in blood, urine, oral and rectal microbiota of renal allograft recipients before and at one- and six-months post-transplant and stated that major changes in the microbiota occurred between pre and one-month post-transplant. We too found that there were significant changes in the relative abundances of the studied taxonomy between pre-and one month-transplant and also observed that there was a significant change in the relative abundance of phyla Bacteroidetes, Firmicutes and Epsilon Proteobacteria between their one and three-months post-transplant period which is in contrary to the report by Fricke et al., where it was stated that the changes persisted post-transplant as there was no significant changes between one and six-months post-transplant. This discrepancy may be due to the changes in sampling time as we did not study the structure six months post-transplant and Fricke et al., haven’t observed the changes at three months. Another factor could be the type of specimen, as we used fresh faecal specimen to characterize gut microbiota and their main focus were to characterize human microbiota which did not include faecal sample.
Our study stands out to be the first in assessing the relationship of pre-transplant gut microbiota with renal allograft rejection. Fricke et al., found an association between pre-transplant human microbiota of oral and rectal samples with post-transplant rejection outcome. Pre-transplant oral samples of 20 patients who never had rejection or any adverse complication post-transplant were compared to five patients who had at least one rejection episode and observed that the Fusobacteria, Proteobacteria and Actinobacteria to be increased in the first group. In rectal samples rejection events in four patients in comparison to 14 patients without rejection showed a decrease in phylum Firmicutes.25 In our study we compared the pre-transplant gut microbiota in faecal samples of 13 recipients who had rejection with 25 recipients who did not have rejection and found that recipients who had a higher relative abundance of phylum Firmicutes before transplantation were more prone to allograft rejection post-transplant, supporting the findings of Fricke et al. To further strengthen our finding, we performed a ROC analysis and found that the ratio of F/B are sensitive and specific indicators of renal allograft rejection, hence if this is further worked extensively it could serve as a valuable strategy to identify patients at a risk of allograft rejection for intense monitoring or alteration of immune suppressive regimen.
Very few studies have found a temporal association of gut microbiota with acute rejection. Ren et al., studied the potential application of gut microbiota as a biomarker for acute rejection on orthotopic liver transplantation in rat models and found that there is an increase in phylum Bacteroidetes and decrease in phylum Firmicutes at the time of rejection.26 Similarly, Oh et al., compared the ileal microbiota in small bowel transplantation and found that there is an increase in phylum Proteobacteria and decrease in phylum Firmicutes at the time of rejection.27 In our study we could not collect samples exactly at the time of biopsy or graft dysfunction, but rejection episodes that occurred nearby our sample collection points (one and three-months post-transplant) were analysed to see the temporal association. We had 10 rejection cases in first three months post-transplant which was compared with 28 non-rejection cases and found that there was a decrease in phylum Firmicutes (not significant) and a significant increase in phylum Bacteroidetes around the time of biopsy proven rejections. Even though our findings with a decrease in the relative abundance of Firmicutes in the rejection group is on par with findings in liver and small bowel transplant, a contradiction occurred with the data represented by Lee et al., where they found a decrease in phylum Bacteroidetes during rejection22 in renal transplant recipients. We postulate this difference to a microbial imbalance as we initially stated that Firmicutes is generally less in post-transplant period in comparison to the pre-transplant period, a further decrease in the Firmicutes post-transplant could also be a risk factor for rejection as either a decrease or increase in the gut flora leads to dysbiosis leading to unfavourable conditions.
The enumeration of microbiota is very much essential and a plethora of methods for addressing it includes cultivation, PCR and sequencing, culturomics28 and hybridization microarrays.29-31 Of these the real-time qPCR method is superior in terms of generally available equipment, cheaper sample preparation, flexibility in application and robustness for diagnostic approaches.32 Various studies33-39 have evaluated the use of phylum and class specific probes to quantify the taxonomy of the predominant gut microbiota which has significant role in disease using real-time PCR. We used qPCR technique to study the predominant occupiers providing a flexible platform for its replication, future use as a potential biomarker and translational utility.
Through this study we have provided useful information on the microbiota changes of the predominant groups in renal transplantation. An extensive study is required to determine the exact role of the observed changes on whether dysbiosis caused complications or whether the complications created the dysbiosis. Our research proved that the pre-transplant levels of F/B ratio serve as an important tool to classify patients at a risk of rejection which is a very valuable finding that needs application in a larger cohort. Thus, the evidence present in this study emphasizes the role of gut microbiota as a diagnostic tool in the earlier detection and prevention of rejection paving way for personalized immune suppressive regimen and improved graft outcome.
ACKNOWLEDGMENTS
The authors would like to thank research participants for their willingness to participate in this research and for the hassle free sample collection and submission without which the work would not have been possible. The authors are also grateful to Jawaharlal Institute of Postgraduate Medical Education and Research (JIPMER) for providing a platform to perform the research.
CONFLICT OF INTEREST
All authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
CP, CPGK, SP and RNG conceived and designed the study. CP and RNG structured the work order. CP conducted experiments. CP and SK collected and analyzed the data. CP wrote the draft manuscript. SP, PV and RNG revised the manuscript. All authors read and approved the final manuscript for publication
FUNDING
The work is funded primarily by Department of Biotechnology, India (DBT e-ProMIS scheme, Approval no. BT/PR24816/MED/30/1902/2017) and Institute’s intramural seed research grant for PhD program dated 2017. The laboratory infrastructure was funded by Department of Science and Technology, India (DST SERB approval no EMR/2016/003382).
DATA AVAILABILITY
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
ETHICS STATEMENT
The study was approved by Institute Ethics Committee Jawaharlal Institute of Postgraduate Medical Education and Research (JIPMER) vide order no. JIP/IEC/2017/0115 dated 25th May 2017.
INFORMED CONSENT
Written informed consent was obtained from the participants before enrolling in the study.
- Shrestha BM. Immune System and Kidney Transplantation. J Nepal Med Assoc. 2017;56(208):482-486.
Crossref - Fishman JA, Thomson AW. Clinical implications of basic science discoveries: Immune homeostasis and the microbiome – Dietary and therapeutic modulation and implications for transplantation. Am J Transplant. 2015;15(7):1755-1758.
Crossref - Maynard CL, Elson CO, Hatton RD, Weaver CT. Reciprocal interactions of the intestinal microbiota and immune system. Nature. 2012;489(7415):231-241.
Crossref - Bartman C, Chong AS, Alegre ML. The influence of the microbiota on the immune response to transplantation. Curr Opin Organ Transplant. 2015;20(1):1-7.
Crossref - Bromberg JS, Fricke WF, Brinkman CC, Simon T, Mongodin EF. Microbiota – Implications for immunity and transplantation. Nat Rev Nephrol. 2015;11(6):342-353.
Crossref - Jandhyala SM, Talukdar R, Subramanyam C, Vuyyuru H, Sasikala M, Reddy DN. Role of the normal gut microbiota. World J Gastroenterol. 2015;21(29):8787.
Crossref - Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59-65.
Crossref - Rinninella E, Raoul P, Cintoni M, et al. What is the Healthy Gut Microbiota Composition? A Changing Ecosystem across Age, Environment, Diet, and Diseases. Microorganisms. 2019;7(1):14.
Crossref - Gibiino G, Lopetuso LR, Scaldaferri F, Rizzatti G, Binda C, Gasbarrini A. Exploring Bacteroidetes: Metabolic key points and immunological tricks of our gut commensals. Dig Liver Dise. 2018;50(7):635-639.
Crossref - Wilkins LJ, Monga M, Miller AW. Defining Dysbiosis for a Cluster of Chronic Diseases. Sci Rep. 2019;9(1).12918.
Crossref - Rossi M, Johnson DW, Campbell KL. The Kidney-Gut Axis: Implications for Nutrition Care. J Ren Nutr. 2015;25(5):399-403.
Crossref - Al Khodor S, Shatat IF. Gut microbiome and kidney disease: a bidirectional relationship. Pediatr Nephrol. 2017;32(6):921-931.
Crossref - Briskey D, Tucker P, Johnson DW, Coombes JS. The role of the gastrointestinal tract and microbiota on uremic toxins and chronic kidney disease development. Clin Exp Nephrol. 2017;21(1):7-15.
Crossref - Wanchai K, Pongchaidecha A, Chatsudthipong V, Chattipakorn SC, Chattipakorn N, Lungkaphin A. Role of Gastrointestinal Microbiota on Kidney Injury and the Obese Condition. Am J Med Sci. 2017;353(1):59-69.
Crossref - Stojanov S, Berlec A, Strukelj B. The influence of probiotics on the firmicutes/ bacteroidetes ratio in the treatment of obesity and inflammatory bowel disease. Microorganisms. 2020;8(11):1-16.
Crossref - Sabatino A, Regolisti G, Brusasco I, Cabassi A, Morabito S, Fiaccadori E. Alterations of intestinal barrier and microbiota in chronic kidney disease. Nephrol Dial Transplant. 2015;30(6):924-933.
Crossref - Roufosse C, Simmonds N, Clahsen-Van Groningen M, et al. A 2018 Reference Guide to the Banff Classification of Renal Allograft Pathology. Transplantation. 2018;102(11):1795-1814.
Crossref - Wu WK, Chen CC, Panyod S, et al. Optimization of fecal sample processing for microbiome study – The journey from bathroom to bench. J Formos Med Assoc. 2019;118(2):545-555.
Crossref - Wesolowska-Andersen A, Bahl MI, Carvalho V, et al. Choice of bacterial DNA extraction method from fecal material influences community structure as evaluated by metagenomic analysis. Microbiome. 2014;2(1).
Crossref - Carroll IM, Ringel-Kulka T, Siddle JP, Klaenhammer TR, Ringel Y. Characterization of the Fecal Microbiota Using High-Throughput Sequencing Reveals a Stable Microbial Community during Storage. PLoS ONE. 2012;7(10):e46953.
Crossref - Li W, Ma ZS. FBA Ecological Guild: Trio of Firmicutes-Bacteroidetes Alliance against Actinobacteria in Human Oral Microbiome. Sci Rep. 2020;10(1):287.
Crossref - Lee JR, Muthukumar T, Dadhania D, et al. Gut microbial community structure and complications after kidney transplantation: A pilot study. Transplantation. 2014;98(7):697-705.
Crossref - Huttenhower C, Gevers D, Knight R, et al. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207-214.
Crossref - Vaziri ND, Wong J, Pahl M, et al. Chronic kidney disease alters intestinal microbial flora. Kidney Int. 2013;83(2):308-315.
Crossref - Fricke WF, Maddox C, Song Y, Bromberg JS. Human microbiota characterization in the course of renal transplantation. Am J Transplant. 2014;14(2):416-427.
Crossref - Ren Z, Jiang J, Lu H, et al. Intestinal microbial variation may predict early acute rejection after liver transplantation in rats. Transplantation. 2014;98(8):844-852.
Crossref - Oh PL, Martinez I, Sun Y, Walter J, Peterson DA, Mercer DF. Characterization of the ileal microbiota in rejecting and non-rejecting recipients of small bowel transplants. Am J Transplant. 2012;12(3):753-762.
Crossref - Lagier JC, Dubourg G, Million M, et al. Culturing the human microbiota and culturomics. Nat Rev Microbiol. 2018;16(9):540-550.
Crossref - Thissen JB, Be NA, McLoughlin K, et al. Axiom Microbiome Array, the next generation microarray for high-throughput pathogen and microbiome analysis. PLoS ONE. 2019;14(2).
Crossref - El Kaoutari A, Armougom F, Leroy Q, et al. Development and validation of a microarray for the investigation of the CAZymes encoded by the human gut microbiome. PLoS ONE. 2013;8(12).
Crossref - Rajilic-Stojanovic M, Heilig HGHJ, Molenaar D, et al. Development and application of the human intestinal tract chip, a phylogenetic microarray: analysis of universally conserved phylotypes in the abundant microbiota of young and elderly adults. Environ Microbiol. 2009;11(7):1736-1751.
Crossref - Kurina I, Popenko A, Klimenko N, et al. Development of qPCR platform with probes for quantifying prevalent and biomedically relevant human gut microbial taxa. Mol Cell Probes. 2020;52:101570.
Crossref - Guo X, Xia X, Tang R, Zhou J, Zhao H, Wang K. Development of a real-time PCR method for Firmicutes and Bacteroidetes in faeces and its application to quantify intestinal population of obese and lean pigs. Lett Appl Microbiol. 2008;47(5):367-373.
Crossref - Yang YW, Chen MK, Yang BY, et al. Use of 16S rRNA gene-targeted group-specific primers for real-time PCR analysis of predominant bacteria in mouse feces. Appl Environ Microbiol. 2015;81(19):6749-6756.
Crossref - Becker AA, Hesta M, Hollants J, Janssens GP, Huys G. Phylogenetic analysis of faecal microbiota from captive cheetahs reveals underrepresentation of Bacteroidetes and Bifidobacteriaceae. BMC Microbiol. 2014;14(1).
Crossref - Cox LM, Cho I, Young SA, et al. The nonfermentable dietary fiber hydroxypropyl methylcellulose modulates intestinal microbiota. The FASEB Journal. 2013;27(2):692-702.
Crossref - Ijssennagger N, Belzer C, Hooiveld GJ, et al. Gut microbiota facilitates dietary heme-induced epithelial hyperproliferation by opening the mucus barrier in colon. Proc Natl Acad Sci U S A. 2015;112(32):10038-10043.
Crossref - Guo X, Xia X, Tang R, Wang K. Real-time PCR quantification of the predominant bacterial divisions in the distal gut of Meishan and Landrace pigs. Anaerobe. 2008;14(4):224-228.
Crossref - Bacchetti De Gregoris T, Aldred N, Clare AS, Burgess JG. Improvement of phylum- and class-specific primers for real-time PCR quantification of bacterial taxa. J MicrobiolMethods. 2011;86(3):351-356.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.