ISSN: 0973-7510
E-ISSN: 2581-690X
Aflatoxins (AFs) are the most potent and ubiquitously found mycotoxins, capable of causing contamination in agricultural products. Aflatoxin B1 (AFB1) is the most toxic and primarily produced Aflatoxin and will be a real threat to the safety of food and feeds. The current study searched for the potential of Lactic acid bacteria (LAB) isolated from animal excreta for AFB1 mitigation. Three LAB out of 56 isolates were found to exhibit more than 50% sorbent action with AFB1 in phosphate-buffered saline (PBS) and were identified as Lactococcuslactis strain CF_6 (OP183481) (65.38%), Lacticaseibacillus casei strain CW_3 (OP183482)(52.63%) and Lactobacillus acidophilus strain CE_4 (OP183483)(63.13%). More than 60% of the total AFB1 removal was observed in 2 hr of incubation, and maximum sorbent action was found at a pH 6-7 range at 37oC for 24 hours. In the Scanning Electron Microscope (SEM) analysis, heat-killed cells showed a significant increase in cell surface binding area, which improved the surface binding for all isolates except L. casei strain CW_3; however, it proves that LAB surface binding is strain-specific rather than heat treatment. Moreover, the rise in AFB1 concentration improved the rate of the sorbent action but did not observe any substantial changes in total AFB1 detoxification. So, it is concluded that the animal excreta may be a versatile source of probiotic LAB for AFB1 detoxification by surface binding.
Aflatoxin B1, Lactic Acid Bacteria, Carcinogen, Sorbent Action, Food Safety, Mycotoxin
Aflatoxins (AFs) are secondary metabolites of the polyketides family, majorly produced by different species of Aspergillus viz., Aspergillus flavus, A. parasiticus and A. nominus, but AFs also reported from various fungi of Aspergillus section, Flavi.1 Strikingly, the causative agents are cosmopolitan distributed endophytic fungi with extreme survivability in unfavourable conditions. AFs are commonly found in most staple foods, spices, oil seeds, tree nuts, milk, meat, eggs and even smoked dry fish and cause life-threatening acute or chronic biological effects in humans and animals.2 The prevalence of AFs in food and feed materials could cause major socio-economic issues in all agriculture-based developing countries from Africa, Latin America and Asia. From the perspective of medical complications, natural AFs have been categorised as the most potent human carcinogens (Group I) by the International Agency for Research on Cancer (IARC), and their exceptional structural stabilities under various food processing conditions make them a potent threat to global food quality. The efficient management of AFs in agri-products requires a massive coordinated approach, which includes technology integration, farmer’s awareness, infrastructure developments and logistics, mandatory implementation and maintenance of legitimate regulations. Moreover, in the era of transcontinental trade, AFs have become a top international food safety threat, and inefficient regulations will severely affect agriculture-based economies and the health of the global population.3
According to the WHO, there are more than 20 AFs have been reported; four among them, Aflatoxin B1(AFB1), Aflatoxin B2(AFB2), Aflatoxin G1(AFG1) and Aflatoxin G2(AFG2) are prominent natural AFs producing severe health effects to humans and animals. Meanwhile, metabolic by-products of AFB1 and AFB2 by the action of the hepatic system of mammals, viz., Aflatoxin M1 (AFM1) and Aflatoxin M2 (AFM2), also exert some threats.4 However, Aflatoxin B1 (AFB1) is the most concerned AFs because of their potential toxicity and substantial production quantity.1 From the structural perspective, AFs are polyketide metabolites with a Difurocoumarolactone structure, which comprises a bifuran ring fused with a coumarin nucleus which is in combination with pentenone or lactone group which varies with the types of AFs.5 Furthermore, in the biological system, AFs act as mutagen, teratogen, immunosuppressive and growth-inhibiting agents,6 and will cause acute or chronic Aflatoxicosis ranging from mild to life-threatening. However, the early reports prove that immunocompetent older adults, children and pregnant women are the prime victims of Aflatoxicosis.
Mitigation of aflatoxin contamination from food or feed could be attained through preventive and degradative ways. The preventive methods are more effective and efficient, which constitute Good Manufacturing Practices (GMP), Good Agricultural Practices (GAP) and standard operating procedures (SOPs) by regulative authorities of global and regional origin for pre-harvesting, postharvesting, packaging, storage and logistics activities of food and feed crops.7 However, solutions for the AFs contaminated crops are tough to accomplish and can be achieved through various detoxification methods viz., physical, chemical and biological nature and among them, biological mode shows more promising, notably the microbial involvement in the mitigation of AFs through metabolisation or reduced bio-availability by sorbent action in the biological system.8
Probiotics are live microorganisms that, when administered in adequate amounts, confer a health benefit on the host (FAO/WHO). Notably, the intervention of probiotics in the AFs mitigation also causes added betterment in a biological system, which points out the importance of research in probiotics-based AFs mitigation.9 The role of Probiotics prominently by Lactic acid bacteria(LAB) and yeast reduce the availability of AFs in a biological system by harnessing their sorbent actions possessed by the specialised cell wall structure with specific functional groups.10 The total sum of adsorption forces exerted by the peptidoglycan, teichoic acid, b-D-glucan and proteins on the cell wall surface with AFs resulted in a conjugate form, which prevents the availability of the toxin in the Gastrointestinal (GI) tract for absorption.11 However, the sorbent action of LAB is reported to bestrain-specific; similarly, the structural changes in the cell wall by heat or cold treatment also affect the adsorption.12 Moreover, Probiotics are need-of-the-hour tools for many biological ailments and have been used in food since the ancient periods onward and possess the status of GRAS (Generally Recognised As Safe) by the FDA and Qualified Presumption of Safety (QPS) status by EFSA (European Food Safety Authority).
This study aims to identify potential probiotic LAB in AFB1 mitigation by using the excreta as the source of probiotics. The versatility of LAB and AFB1 degradation properties will make an optimum combination and promote the use of AFs contaminated agriculture products prominently in the form of fermented foods. For this study, different samples, including Cow dung, Chicken excreta and Calf faeces, were collected as the source of LAB.
Isolation and preliminary characterisation of LAB strains
Cow dung, calf faeces of 15 days of age and Chicken droppings were selected from the places around Gandhigram, Dindigul, India, for probiotic isolation. About 1 gm of the sample was serially diluted up to 10-7 times in distilled water; 0.1ml from the dilutions of 10-5, 10-6and 10-7 were spread plated in MRS Agar (DeMan, Rogosa and Sharpe Agar), and incubated under anaerobic conditions for 48-72 hours at 37°C in an anaerobic jar filled with CO213 the well-developed, distinct and creamy white characterised colonies were selected and stored for further screening and identification. Then, the selected isolates were checked for cell morphology by gram staining spore-forming and motility; a catalase test was also performed. The gram-positive and catalase-negative isolates with characteristic morphological properties were selected and stored in MRS Agar at -4°C for further screening and characterisation of AFB1 removal by adsorption.
Screening for AFB1 binding isolates
AFB1 binding assay
AFB1 stock was prepared by making up 1mg of AFB1(Sigma-Aldrich) into 50ml with methanol, and working standards of 2,4 and 8 mgL-1 concentration were prepared from the stock by making up with methanol and 1% acetic acid in a 1:1 ratio.
AFB1 binding assay was carried out by modified AOAC 991.31 method with Immunoaffinity column (IAC) cleanup(Afla B, Vicam) followed by HPLC quantification. HPLC system with a post-column derivatisation system by an electrochemical cell (KOBRA® Cell) and fluorescent detector (FLD) was used.
50 ml of LAB cultures in MRS broth were incubated with shaking at 37°C for 24 hrs.14 Cells were collected by centrifugation at 8000 rpm for 20 min, and the cultural pellets were washed in Phosphate-buffer saline (PBS) of pH 7.2 two times. Finally, bacterial pellets were re-suspended in 5 ml of sterile PBS and cells were adjusted to 109 cells/ml concentration by OD600. One millilitre (1 ml) of cell suspension was centrifuged at 8000 rpm for 10 min, and the supernatant was removed completely. To the cell pellet, 950 µl of PBS (pH 7) was added along with 50 µl of AFB1(Sigma-Aldrich) of 2 mgL-1 and mixed thoroughly and incubated at 37°C at 120 rpm with an incubation shaker under dark conditions up to 48hr of incubation time. 950 µl of PBS (pH 7) and 50 µl of AFB1 of 2 mgL-1 standards were used as a control. After incubation, the supernatant was removed by centrifugation at 10,000 rpm for 10 min, and the pelleted bacteria were also subjected to two times wash with 1ml PBS; the primary supernatant and washed supernatant was collected and subjected to Immunoaffinity column (IAC) cleanup (Afla B, Vicam) at the rate of one drop per second followed by a two-time wash with ultrapure water. The AFB1 from the IAC was extracted by 1 ml HPLC grade methanol and was subjected to quantification by the HPLC-FLD system.
Quantification of AFB1
For the quantification of AFB1, a reverse phase HPLC system (Shimadzu, Tokyo, Japan) consisted of a gradient pump (LC-20AT), a C18 column (250mm x 4.6mm, 5µm Shiseido, Japan) and a Fluorescent detector with a post-column derivatisation Electrochemical cell (KOBRA® Cell) equipment was used. 20µl of the cleaned-up sample was injected, where micro-filtered methanol-water (40:60 v/v) with a trace amount of KBr and 4M HNO3 (400ml HPLC methanol+600ml HPLC Water+119 mg KBr+ 350µl 4 M HNO3) was used as mobile phase with a flow rate of 1.0 ml/ min at 35°C. AFB1 detection was accomplished by a Fluorescent detector (Excitation: 365 nm, Emission: 455 nm). The retention time of AFB1 was approximately 16.39 min. The percentage of AFB1 removed by the bacterial suspension was calculated using the formula.
%AFB1 = [ 1-AFB1 peak area of the sample / AFB1 peak area of toxin control ] × 100
Validation of AFB1 HPLC-FLD method
Method validation was carried out using five different concentrations of AFB1 (20,50, 100,200, &500 µgL-1) based on Muscarella et al. (2009)41 method. The LOD (Limit of Detection) and LOQ (Limit of Quantification) were calculated by using the formula LOQ = 10 x σ/m and LOD = 3.3 x σ/m, where σ = residual standard deviation and m = slope of the calibration curve.15 Similarly, the recovery was carried out in three different concentrations of AFB1 spiked phosphate-buffered saline (PBS) (pH 7.2), and recovery was calculated using the formula
Recovery (%) = (AFB1 quantity identified/ AFB1 theoretical quantity) × 100.
Biochemical analysis and probiotic characterisation
The selected three LAB isolates were subjected to biochemical analysis based on the procedures by Bergey’s Manual of Systematic Bacteriology.16
Probiotic characterisation was carried out for tolerance against low pH and bile salt. Similarly, the hemolytic property was also checked in the blood agar.
pH tolerance
For the low pH tolerance, pH 1.5 and 3 were selected, which were maintained by hydrochloric acid (1M). 1% (v/v) of overnight cultured inoculum in MRS broth with corresponding pH was selected, and incubation was carried out for 11 hrs anaerobically at 37°C, where standard MRS broth was used as the control.17 The pH resistance potential was identified in percentage (%) by the standard plate count method (CFU/ml), and the experiment was carried out in triplicate.
Survival rate (%) = Viable cell count at specific pH (1.5 & 3)/ Viable cell count in control X 100
Bile salt tolerance
0.3% and 1% of bile salt (Himedia, India) were used to check the bile salt tolerance of three bacterial isolates. In the experiment, 1% (v/v) of inoculum in MRS broth with 0.3% and 1% of bile salt were incubated for 9 hrs under 37 °C in triplicate. The same experimental culture without bile salt acted as the control.18 Determination of survival rate was identified by the standard plated count method (CFU/ml).
Survival rate (%) = Viable cell count at specific bile salt (0.3% & 1%)/ Viable cell count of control X 100
Hemolytic property
The Hemolytic property determined the pathogenicity of the bacterial isolate and was performed in a fresh blood agar plate. 5% of sheep blood is the major constituent of the agar plate and incubated for 48 hrs at 37°C.19 The various types of hemolysis, viz. β-hemolysis (the clear area around the colony), α-hemolysis (green-hued area), and γ-hemolysis (no hemolysis) determine the pathogenicity.
Antibiotic sensitivity
Antibiotic resistance of the selected LAB isolates by Kirby-Bauer disk diffusion method. The antibiotic disc of 6 mm was used, viz. Amikacin (AMK), Gentamycin (GEN), Cefotaxime (CTX) and Penicillin (PEN) were obtained from HiMedia, India. The activated isolates were seeded on the Mueller Hinton Agar, and the corresponding antibiotic discs were placed on the surface. After 24 hrs of incubation at 37°C, the sensitivity of the isolates was checked by measuring the zone of inhibition in diameters around the antibiotic disc.20
Scanning electron microscopy (SEM) analysis
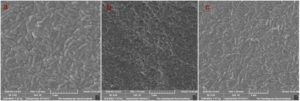
SEM analysis was used to study the morphological characterisation of the isolated LAB and was carried out by the procedure of Oslan et al.21 The overnight-grown cells in MRS media were used for SEM analysis. For the specimen preparation, the bacterial sample was fixed by 2.5%(v/v) glutaraldehyde buffer for 4 hrs at 4°C. This is followed by three times washing with 0.1M sodium cacodylate buffer for 10 min. As a post-fixation, the specimen was treated with 1% (w/v) osmium tetroxide for 2 hrs at 4°C; prior to the dehydration procedure, the sample was washed twice. Acetone at 35-100% was used for the dehydration of the sample, followed by the specimens subjected to dry. Before the SEM analysis, the dried sample was mounted on a carbon-coated stub and got a thin layer of gold from a sputter coater (SC7620, Quorum, United Kingdom) and the microscopy was carried out in VEGA3, TESCAN, Czech Republic.
Molecular identification and phylogenetic analysis of the most potent AFB1 binding LAB
The identification of the three most potent AFB1 binding LAB was confirmed by 16S rRNA sequencing, by the amplification of the 16S rDNA of 16S rRNA gene using universal primers of 27F(5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′) in ProFlexTM2xFlat PCR thermocycler of Applied Biosystems™. The sequenced lactic acid bacteria were identified using NCBI BLAST, USA (Basic Local Alignment Search Tool) similarity search tool and the sequences were submitted to GenBank®. Phylogenetic analysis was performed using the software MEGA 11 by the neighbour-joining method with a scale bar of 0.05.
Characterisation of AFB1 binding by LAB
Sorbent action-based removal of the AFB1 by LAB is one of the extremely complicated modes of detoxification, which has been influenced by a multitude of factors comprising incubation time, pH, concentration of AFB1, microbial load used, viability of LAB and bacterial strains.22 Therefore, the AFB1 binding potential of selected LAB was characterised under specific parameters imperative for AFB1 removal.
Live and heat-inactivated LAB in AFB1 binding
The sorbent action and removal of AFB1 by three potential LAB at live and thermally inactivated stages were carried out. For that, the thermal inactivation was carried out in boiling water for 1 hour and bacterial mortality was confirmed by the overnight incubation in MRS media. The quantification was carried out by the methods mentioned in the AFB1 binding assay.
Effect of pH and temperature in AFB1 binding by LAB
The role of pH in the AFB1 binding with the LAB was carried out at pH 2-8 and accomplished by citrate and phosphate buffer. Similarly, two temperatures were selected for the temperature study, viz., 30 and 37°C.
Effect of AFB1 concentrations in binding by LAB
Three different concentrations of AFB1 were tested against the potent LAB to determine the kinetics of the sorbent action between LAB cells and AFB1. The concentrations of AFB1 tested were 100 µgL-1,200 µgL-1, and 400 µgL-1 in the 1 ml reaction mixture, prepared from working standards of 2, 4and 8 mgL-1 of AFB1, respectively. However, the experiment was carried out for both live and heat-killed LAB at 30 and 37°C 2hr, 12hr, 24hr and 48hr of incubation.
Scanning electron microscopy (SEM) for surface characterisation of AFB1 bound LAB
The morphological changes associated with the selected LAB at live and heat-killed stages with AFB1 treatment and without treatment were analysed by scanning electron microscope (SEM) (VEGA3, TESCAN, Czech Republic). Scanning electron microscopy is a highly useful tool for studying the morphological changes of the bacterial surface at the minute level. The surface characterisation by SEM analysis of the live and heat-killed LAB with or without AFB1 was carried out by Oslan et al.21 proposed method. 109 CFU/ml of LAB with different treatments (live and heat-killed) in 1ml PBS with 100 µgL-1 of AFB1 was incubated for 24 hrs at 37°C. The specimen preparation and SEM analysis were carried out based on the procedure in the Scanning electron microscopy (SEM) analysis section.
Statistical analysis
All the values used in the analysis are the mean value of independent three-time replicated results with standard deviation (Mean±SD) and the significance level tested by ANOVA (Analysis of variance) by using the SPSS statistical tool. The graphical representation was carried out in Origin 8.5 software.
Isolation and preliminary characterisation of LAB
A total of 56 presumptive bacterial isolates were selected from the MRS media. All were gram-positive and catalase-negative with characteristic morphological appearances like cream to off-white colour smooth colonies. A prominent portion of LAB isolate was sourced mainly from Cow dung, with 26 isolates, followed by Chicken excreta and Calf faeces, with 22 and 8 isolates, respectively.
HPLC-FLD method validation
The calibration curve was prepared from the values of five different concentrations of AFB1(20, 50, 100, 200 & 500 µgL-1), found linear, and its coefficient of determination (R2) value was found to be satisfactory (0.999) (Table 1). The Limit of Detection (LOD) and Limit of Quantification (LOQ) were identified as 0. 65 and 1.98µgL-1, respectively and found to be aligned with the AFB1 limit by the statutory authorities. Similarly, the overall recovery of AFB1 in PBS was identified as 95.79% for three different concentrations of AFB1. Meantime, the % RSD (Relative standard deviation) was found in the range of (0.51%-1.32%). Because of the safe overall recovery and % RSD, the method validity was found satisfactory, the retention time (RT) of AFB1 was found at 16.40 minutes, and any interfering peaks were not observed in the chromatogram.
Table (1):
HPLC-FLD method Linearity, sensitivity and recovery of AFB1 in PBS
| Linearity and sensitivity* | ||||||
|---|---|---|---|---|---|---|
| Analyte | Range, µgL-1 | slope | Intercept | R2 | LOD µgL-1 | LOQ µgL-1 |
| AFB1 | 20-500 | 35907 | 3630.8 | 0.9999 | 0.655 | 1.98 |
| Recovery** | ||||||
| Analyte | Spiking concentrations µgL-1 | Recovery % | %RSD | |||
| AFB1 | 5 | 96.18 | 0.92% | |||
| 10 | 96.14 | 1.32% | ||||
| 20 | 95.05 | 0.51% | ||||
*A total of 30 values of five different concentrations were selected to find out the Linearity and sensitivity
**Triplicate values of each spiked concentration were used to identify recovery
Screening for AFB1 binding isolates
25 isolates out of 56 exhibit binding with the toxin at an average minimum of 5% (Table 2), and the binding rate varies with the isolates. There were only three isolates were found to remove above 50% of AFB1 with a maximum of 64.84% (CF6) followed by 62.23% (CE4) and 52.63% (CW3) at 37oC in 24 hours. Furthermore, the AFB1 binding of the remaining 31 isolates was found to be none or negligible. The potential top three isolates, viz., CF6, CE4 and CW3, were selected to identify and characterise the AFB1 binding. As per the earlier studies, Lactic acid bacteria-based AFB1 removal has been accomplished mainly through surface binding with microbial cell walls10 and forced to reduce the bio-availability of the toxin in the biological system.
Table (2):
List of LAB isolates and their AFB1 detoxification potentials
No. |
Source |
Isolate identification |
AFB1 binding in % |
|---|---|---|---|
1 |
Cow dung |
CW5 |
5.23±0.95 |
2 |
Chicken excreta |
CE18 |
5.68±1.01 |
3 |
Cow dung |
CW15 |
5.72±0.84 |
4 |
Cow dung |
CW12 |
6.89±0.94 |
5 |
Cow dung |
CW7 |
7.14±1.07 |
6 |
Chicken excreta |
CE4 |
7.86±1.24 |
7 |
Cow dung |
CW6 |
7.91±0.86 |
8 |
Chicken excreta |
CE10 |
8.17±0.749 |
9 |
Chicken excreta |
CE13 |
9.14±0.943 |
10 |
Cow dung |
CW22. |
9.89±0.34 |
11 |
Cow dung |
CW9 |
10.48±1.16 |
12 |
Chicken excreta |
CE2 |
10.67±0.37 |
13 |
Calf faeces |
CF2 |
11.64±0.94 |
14 |
Cow dung |
CW13 |
11.49±0.81 |
15 |
Cow dung |
CW18 |
13.48±1.04 |
16 |
Cow dung |
CW4 |
15.49±1.67 |
17 |
Cow dung |
CW6 |
15.73±0.64 |
18 |
Chicken excreta |
CE9 |
16.84±0.91 |
19 |
Chicken excreta |
CE11 |
17.59±1.27 |
20 |
Calf faeces |
CF1 |
17.61±0.81 |
21 |
Calf faeces |
CF4 |
19.48±1.07 |
22 |
Chicken excreta |
CE17 |
19.61±0.59 |
23 |
Cow dung |
CW3(OP183482) |
52.64±0.89 |
24 |
Chicken excreta |
CE4(OP183483) |
62.23±0.71 |
25 |
Calf faeces |
CF6(OP183481) |
64.84 ±0.94 |
*Values represented in percentage as a Mean ± Standard deviation of three-time replicated values.
Biochemical analysis and probiotic characterisation
The biochemical analysis performed by the three selected isolates is presented in Table 3. In probiotic characterisation, isolate CW3 showed the most tolerance at pH 1.5, followed by CE4 and CF6; at pH 3, the viabilities of all three isolates were found to be more favourable. Meanwhile, more than 51% viability was observed in 0.3% of bile salt by all the isolates; among them, isolate CF6 was identified as the most promising, and the detailed results are presented in Table 4. Both the parameters were found to be statistically significant (p ≤ 0.05). While checking the pathogenicity in the blood agar, none of the isolates produced hemolysis in the media. While doing the antibiotic sensitivity test, none of the isolates showed resistance to the selected antibiotics, as presented in Table 5. Meanwhile, in the SEM image of the isolates, as presented in Figure 1, both the isolate CE4 and CW3 had a characteristic rod-shaped morphology, while the isolate CF6 produced a cocci structure.
Table (3):
Morphological and biochemical characteristics of isolated LAB
Tests |
Lactococcus lactis CF_6 |
Lacticaseibacillus casei CW_3 |
Lactobacillus acidophilus CE_4 |
|---|---|---|---|
Colony Characteristics in MRS Media |
Creamy white smooth colony |
Creamy white smooth colony |
Creamy white circular colony |
Morphology Of Bacterial Cells |
Cocci |
Rod |
Rod |
Gram Staining |
+ |
+ |
+ |
Motility |
Non-motile |
Non-motile |
Non-motile |
Spore Producing |
Non-spore forming |
Non-spore forming |
Non-spore forming |
Indole Production Test |
– |
– |
– |
Oxidase Test |
– |
– |
– |
Catalase Test |
– |
– |
– |
Nitrate Reduction Test |
– |
– |
– |
Citrate Utilisation Test |
– |
+ |
– |
Methyl Red Test(MR) |
+ |
– |
+ |
Voges Proskauer Test(VP) |
+ |
– |
– |
Starch hydrolysis |
– |
+ |
+ |
Casein hydrolysis |
+ |
+ |
+ |
Hemolysis Property |
Non-hemolytic |
Non-hemolytic |
Non-hemolytic |
Carbohydrate Fermentation |
|||
Maltose |
+ |
+ |
+ |
Galactose |
+ |
+ |
+ |
Sucrose |
+ |
+ |
+ |
Fructose |
+ |
+ |
+ |
Lactose |
+ |
+ |
+ |
*- represents negative responses and + represents positive responses
Table (4):
Viability of isolated LAB isolate under various pH and Bile salt concentrations at 37°C
| Isolate | Survival rate (%) at various pH in MRS broth | Survival rate (%) at various concentrations of Bile salt in MRS broth | ||
|---|---|---|---|---|
| CF6 | pH 1.5 | pH 3 | 0.3 % | 1 % |
| 42.47 ± 1.81 | 85.91 ± 1.90 | 67.58 ± 3.62 | 19.38 ± 2.70 | |
| CW3 | 50.24 ± 1.65 | 91.01 ± 3.07 | 51.98 ± 1.60 | 7.04 ± 1.12 |
| CE4 | 44.38 ± 2.88 | 81.69 ± 3.93 | 57.85 ± 3.42 | 14.46 ± 2.18 |
* Values represented in percentage as a Mean ± Standard deviation of three-time replicated values.
Table (5):
Antibiotic sensitivity profile of isolated LAB
| No. | Antibiotics | LAB isolates | ||
|---|---|---|---|---|
| L. lactis CF6 | L.casei CW3 | L.acidophilus CE4 | ||
| 1 | Penicillin (PEN) | 51.6 ± 0.57 | 18.3 ± 0.62 | 55.3 ± 0.57 |
| 2 | Gentamycin (GEN) | 27.3 ± 0.53 | 26.6 ± 0.57 | 27 ± 1 |
| 3 | Amikacin (AMK) | 26 ± 0.31 | 30 ± 1 | 30.3 ± 0.61 |
| 4 | Cefotaxime (CTX) | 40.3 ± 0.47 | 23.3 ± 0.63 | 59.6 ± 1.15 |
* Values represented in millimetres (mm) as a Mean ± Standard deviation of three-time replicated values.
Molecular identification and phylogenetic analysis
Three selected LAB isolates were subjected to molecular sequencing of 16S rRNA and were identified by NCBI BLAST, USA and the sequences were submitted to GenBank. The GenBank accession number and their identities are presented in Table 6. Then, the kinship relationship of the isolates was determined by a phylogenetic tree created by MEGA 11 software and is presented in Figure 2. The isolate CF6 showed a similarity of 98.87% at 99% of query cover with Lactococcus lactis strain CJNU 3001, while the isolate CW3 showed maximum similarity with Lacticaseibacillus casei strain NCDO 161 at a similarity of 92.11% with 99% of query cover and the isolate CE4 found 97.72% of similarities at 99% of query cover with Lactobacillus acidophilus strain EMBS081. There were reports of AFB1 sorbent potential by L. lactis(KC834394) isolated from fermented food by Singh et al.14 with a high AFB1 binding property (74.56%); similarly, 27% of AFB1 was reported by Sezer et al.23 AFB1 binding by L.caseiwas also reported earlier. Liew et al.11 reported 98% of sorbent action by a specific strain of L. casei Shirota (Lcs). In another work by Pizzolitto et al.24 mentioned 27.6 % of AFB1 removal by L. casei 1. The sorbent action of L. acidophilus with AFB1 was also reported by Marrez et al.,25 and 80% of AFB1 reduction was attained.
Table (6):
Details of molecular identification of the potential isolates with NCBI Accession
No. |
Isolate |
Identified organism |
GenBank Accession number, NCBI |
|---|---|---|---|
1 |
CF6 |
Lactococcus lactis strain CF_6 |
OP183481 |
2 |
CW3 |
Lacticaseibacillus casei strain CW_3 |
OP183482 |
3 |
CE4 |
Lactobacillus acidophilus strain CE_4 |
OP183483 |
Characterisation of AFB1 binding LAB
Live and heat-killed microbial cells in AFB1 binding
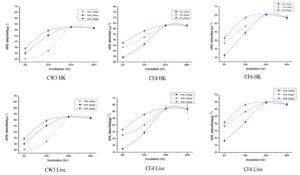
As shown in Figure 3, two heat-killed LABs, CF6 (L. lactis) and CE4 (L. acidophilus), showed more sorbent removal of the AFB1. In contrast, isolate CW3 (L. casei.) showed opposite results, in which live cells produce slightly more AFB1 removal (Table 7). However, the sorbent action of LAB in the heat-killed stage confirms AFB1 removal by the surface action. It can exclude the involvement of any other metabolic mechanism in the removal of AB1. The isolate CF6 produced a maximum AFB1removal of 65.31%, followed by CE4 with 62.64%, and CW3 showed removal efficiency of 52.61% at 37°C for 24 hours under heat killed stage. Furthermore, it reveals that the AFB1removing the potential of the heat-treated LAB is strain-dependent. Moreover, statistical significance (p ≤ 0.05) was observed for live and heat-killed LAB in AFB1 removal.
Table (7):
AFB1 removal (in %) by three LAB isolates in Live and Heat Killed (HK) state at two different temperature and pH 7 in various incubation time
| At 37oC | Incubation time in hours | |||||||
|---|---|---|---|---|---|---|---|---|
| 2 hours | 12 hours | 24 hours | 48 hours | |||||
| HK** | Live | HK | Live | HK | Live | HK | Live | |
| L. lactis CF6 | 41.29 ± 0.85* | 38.19 ± 1.34 | 54.49 ± 0.54 | 51.28 ± 0.67 | 65.31 ± 0.75 | 64.84 ± 0.83 | 63.58 ± 0.49 | 63.4 ± 0.11 |
| L. acidophilus CE4 | 34.28 ± 0.36 | 32.54 ± 0.14 | 46.21 ± 0.33 | 44.46 ± 0.203 | 63.00 ± 0.51 | 62.22 ± 0.75 | 63.87 ± 1.14 | 62.74 ± 0.91 |
| L. casei CW3 | 25.34 ± 0.43 | 26.34 ± 0.64 | 32.24 ± 0.32 | 33.22 ± 0.84 | 52.61 ± 0.24 | 53.14 ± 0.13 | 51.81 ± 0.67 | 52.52 ± 0.92 |
| At 30oC | ||||||||
| L. lactis CF6 | 39.52 ± 0.85 | 38.17 ± 1.18 | 52.54 ± 0.94 | 51.05 ± 0.59 | 64.61 ± 0.79 | 63.85 ± 0.91 | 63.81 ± 1.06 | 63.44 ± 1.07 |
| L. acidophilus CE4 | 34.06 ± 0.93 | 32.38 ± 0.81 | 45.82 ± 0.82 | 44.36 ± 0.78 | 61.32 ± 0.62 | 60.19 ± 1.16 | 61.36 ± 0.95 | 60.48 ± 1.08 |
| L. casei CW3 | 23.56 ± 1.06 | 24.53 ± 0.71 | 31.05 ± 0.61 | 31.93 ± 0.72 | 51.88 ± 1.04 | 52.41 ± 0.92 | 51.04 ± 1.09 | 51.94 ± 1.17 |
*Results represented are the Mean of three replicate values in percentage with Standard deviation
** K represents Heat-Killed LAB cells
Figure 3. AFB1 detoxification in percentage by both live and heat-killed LAB isolates at 30 and 37°C and pH7 for 24 hrs of incubation. Values are represented as the Mean of three replicates with standard deviation, where HK stands for heat-killed
Temperature in AFB1 binding
All three selected strains of LAB could remove AFB1 in the liquid media at two distinct temperatures at 30 and 37°C (Table 7). However, the temperature at 37oC was the most optimum for AFB1 sorbent action by LAB at both live and heat-killed stages. The isolate CF6 ( L. lactis) was found to produce maximum potential at 37°C; however, the LAB at 30°C also showed a substantial quantity of AFB1 detoxification as compared with the optimum AFB1 removal at 37°C. Therefore, the optimum detoxification at 37°C proves their role in fermented food for oral consumption.
Table (8):
AFB1 removal (%) by live and heat-treated isolates under various pH at 37°C for 24 hours of incubation
Isolates |
pH 2 |
pH 3 |
pH 4 |
pH 5 |
pH 6 |
pH 7 |
pH 8 |
|---|---|---|---|---|---|---|---|
(HK) L. lactis CF6 |
34.43 ± 1.06 |
51.37 ± 1.07 |
59.20 ± 0.85 |
63.20 ± 0.94 |
64.84 ± 0.54 |
65.39 ± 1.03 |
64.17± 0.84 |
(Live) L. lactis CF6 |
32.54 ± 1.07 |
49.47 ± 0.89 |
58.27 ± 0.76 |
60.29 ± 2.05 |
64.11 ± 0.97 |
64.67 ± 0.61 |
64.33 ± 1.17 |
(HK) L.acidophilus |
47.69 ± 1.15 |
51.29 ± 2.04 |
55.86 ± 1.04 |
60.67 ± 0.98 |
61.93 ± 1.07 |
63.13 ± 0.67 |
59.89 ± 0.74 |
(Live) L.acidophilus |
48.31 ± 1.06 |
52.26 ± 1.14 |
55.72 ± 2.07 |
59.41 ± 0.95 |
61.34 ± 0.87 |
62.28 ± 0.79 |
56.37 ± 1.10 |
(HK) L. casei |
42.33 ± 0.95 |
45.31 ± 1.06 |
49.04 ± 1.10 |
50.3 ± 0.85 |
52.78 ± 1.08 |
52.61 ± 1.24 |
52.28 ± 0.94 |
(Live) L. casei |
42.48 ± 1.11 |
46.36 ± 1.06 |
49.87 ± 0.88 |
51.29 ± 0.74 |
53.36 ± 1.13 |
53.14 ± 1.13 |
52.89 ± 0.66 |
*Results represented are Mean of three-time replicate values in percentage with Standard deviation
**HK stands for heat-killed
pH in AFB1 binding
The results as shown in Table 8, the pH 7 was identified as optimum for CF6 ( L. lactis) and CE4 (L.acidophilus) at 37°C for 24 hours of incubation for both live and heat-killed cells, while pH 6 was found to be optimum for CW3 (L. casei.). However, ample detoxification was observed in a pH 5-8 range. The least detoxification was found in the acidic pH 2, and a gradual rise of detoxification was observed up to pH7 for CF6 (L. lactis) and CE4 (L. acidophilus) and started to decline. The detoxification activities are presented in Table 8. This report contrasted with Rahaie et al.4 study, which supports the acidic pH for more surface binding than neutral pH. pH is one of the prominent determinants determining the specific ionic state of the functional groups of the LAB cell wall intended to take part in the sorbent action, resulting in the conjugated compound with AFB1.
In this study, the optimum pH is confined to the neutral pH for most of the LAB isolates; meantime, a significant amount of AFB1 was also removed in the slightly acidic pH (5-6), and the results also showed statistical significance (p< 0.05). However, this result is converse to the report of Kumara et al.26; their results revealed that pH 2 is optimum for binding L. fermentum with the AFB1. Moller et al.27 stated that pH 3 to 6.5 was the optimum pH for the binding mechanism, which varies with different bacteria. At the same time, the study by Singh et al.14 complies with the current results, which reported pH 7 as optimum in their research.
Concentration of AFB1 standard in AFB1 binding
Different concentrations viz., 100 µgL-1, 200 µgL-1 and 400 µgL-1 of AFB1 revealed the pattern of sorbent action with the selected LAB strains. A rise in AFB1 detoxification was observed with the increase in AFB1 concentration in minimum incubation time, and the results are illustrated in Figure 4. However, the increase in AFB1 concentration doesn’t cause any changes in the total aflatoxin detoxification. After reaching a saturation point between AFB1 and LAB, further detoxification was found to resist. The optimum detoxification was observed at 37 °C for 24 hours of incubation at pH 7 for both heat-killed and live LAB isolates; similar results were reported by Singh et al.14 Furthermore, the results were statistically significant (p ≤ 0.05).
Figure 4. Detoxification at various concentrations of AFB1 by heat-killed (HK) and live three LAB isolates (CW3, CE4 and CF6) at 37°C and pH7 up to 48 hr of incubation. Values are represented in µgL-1 as the average of three replicates with standard deviation.
Incubation periods in the AFB1 binding with the LAB
During the different duration of incubation time viz., 2hrs, 12hrs, 24hrs and 48hrs incubation, 24hrs was identified as optimum for selected LAB CF6 ( L. lactis) and CW3 (L. casei.) in AFB1 detoxification, while CE4(L. acidophilus) shown optimum detoxification at 48hrs of incubation (Table 5). However, an average of more than 60% of the total AFB1 detoxification was observed (Table 7) in the minimum incubation of 2hrs, which proves the speedy mechanism of detoxification by surface action.
Scanning electron microscopy (SEM) for surface characterisation of AFB1 bounded LAB
SEM technology is best for studying the morphological changes associated with bacterial cells. In the analysis, the HK cells produced enlarged cells with some structural disintegration compared to the live cells, illustrated in Figure 5. The live cells were more compact and maintained structural integrity, while the HK cells were large and disintegrated. In the SEM analysis, any conformational changes on the LAB cell surface associated with AFB1 binding were not observed, in contrast to the Liew et al.11 report.
A substantial amount of AFB1 binding LAB from various animal excreta has proven its importance as a potential source of LAB, like conventionally accepted dairy products. The early documented studies reported the potential of excreta for isolating LAB.27-30 The identified optimum AFB1 binding LAB have reported extensive use in fermented foods, especially in cereal-based fermented foods,31,14 which are popular in developing countries where most AFs-based hardship occurs,32 according to Badji et al.33 reported 90% AFB1 reduction in plant-based fermented product by L. plantarum. The probiotic characterisation of the isolated LAB proved their potential as safe probiotics. Moreover, these results infer their application in cereal-based fermented food or feed that can be capable of debilitating AFB1-induced biological deleterious effects by reducing the biological availability of AFB1 by the surface binding of LAB in the biological system.
The optimum conditions identified for the potent isolates for AFB1 detoxification backing their role in the cereal-based fermented food, promoting their oral use. Specifically, the temperature for optimum removal of AFB1 was identified in the range of body temperature, which promotes the use of the probiotic bacteria for oral use. According to Singh et al.14 the concentration of the AFs standards is directly proportional to the surface binding up to a saturation point; similarly, 24 hours was found to produce maximum adsorption. However, the minimum incubation period of 2 hours produces more than 50% of the total binding, implying the swift binding action between the LAB cell surface and AFB1. The maximum concentration tested, 400 µgL-1 of AFB1, showed a high binding rate compared with the remaining two concentrations of 100µgL-1 and 200µgL-1.
The SEM analysis for the morphological changes in the bacterial cell surface proved the enlarged size of the heat-treated LAB, which improves the surface binding by providing more surface area. Therefore, it can be interpreted that the HK cells provide more surface area by protruding out the cell surface structural compounds like proteins, teichoic acids and polysaccharides,32-34 similarly exposing hydrophobic pockets that actively participate in the binding of AFs. As per the theoretical compliance, two out of three LAB (CF6 and CE4) isolate was found to increase the surface binding in the heat-treated condition. The prominent heat-stable cell wall compound teichoic acid35 and exposed hydrophobic pockets are presumed to be the reason for the enhanced surface binding for AFB1. Conversely, the HK CW3 cells produced less surface binding than the live cells. As an observation, the induced binding of AFB1 by LAB through heat treatment is believed to be strain-specific.
A slight reduction of AFB1 detoxification was found after 24 hours of incubation for CF6 ( L. lactis) and CW3 (L. casei.). Earlier studies also reported the chances of the reversible nature of the cell-toxin conjugate of AFB1 and LAB cells either in the live or killed stages.36,37 This reversible nature of the sorbent action is believed to be due to the involvement of the lion portion of the weak non-covalent forces in the conjugate formation 22 like hydrophobic, van der Walls forces, electrostatic attractions, etc. and that are also presumed that old cells reduce the sorbent action.14 In our study, the two-time washing of LAB cell pellets with 1ml PBS during AFB1 quantification confirms the strong surface binding between LAB cells and AFB1. However, the reversible nature observed was only a minute fraction of the total AFB1 removed in our study.
In the surface binding mechanism for the removal of AFB1, the viability of the LAB was found to play a major role because the majority of heat-inactivated LAB cells showed high efficiency in the AFB1 removal, and it was also reported by Banwo et al.38 According to Haskard et al.22 and Peltonen et al.,39 the surface binding phenomenon of AFs with the cell surface of LAB is the involvement of the non-covalent interactions with the specific functional groups that are exposed from the surface of the gram-positive cell wall. Also, some hydrophobic pockets are also involved in the binding.40 Therefore, the activity of heat-treated cells is probably due to the exposure of cell wall compounds by the denaturation caused by the heat treatment.41 Similarly, Oluwafemi and Da-Silva42 reported a higher efficiency of heat-killed LAB in removing AFs than live cells, and Ondiek et al.40 also reported increased AFB1 detoxification with heat-treated LAB. Moreover, LAB is also extensively used for the preventive mode of AFs detoxification in the role of an anti-Aspergillus fungi agent. Marlida et al.43 and Asurmendi et al.44 recently reported LAB’s inhibition potential against Aspergillus fungi.
In this study, three lactic acid bacteria (LAB) isolated from animal excreta showed sorbent action with AFB1 and removed a substantial amount of AFB1. Such probiotic microbes will be capable of preventing the biological availability of AFs in the GI tract of the animals. Strikingly, the optimum pH and temperature for detoxification were found at 37oC and neutral to slightly acidic pH. Similarly, The identified LAB are proven food fermenting bacteria with multitudes of biological activities, so this may be used as a mother culture for cereal-based fermented food, which will ameliorate AFB1 toxicity and promote AFs contaminated agriculture products for human as well as animal consumption. Moreover, this study proved that the excreta will be a versatile source of potential LAB for specific biological activities. In conclusion, this study may promote the use of AFB1-contaminated agricultural products by the involvement of LAB without causing drastic health effects. At the same time, a proportionate reduction of contaminated agricultural products in landfills can also be attained, which is considered a major greenhouse gas source. So, this study may be impacted on multidimensional levels rather than health and economic well-being alone.
ACKNOWLEDGMENTS
The authors would like to thank Gandhigram Rural Institute (Deemed to be University) for providing the facilities to conduct this work.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- Frisvad JC, Hubka V, Ezekiel CN, et al. Taxonomy of Aspergillus section Flavi and their production of aflatoxins, ochratoxins and other mycotoxins. Stud Mycol. 2019;93:1-63.
Crossref - Singh IS, Nsokolo E. Prevalence of Aflatoxins in Smoked-Dried and Fresh Fish in Zambia. J Environ Prot. 2020;11(01):13-21.
Crossref - Wu F, Guclu H. Aflatoxin Regulations in a Network of Global Maize Trade. PLoS ONE. 2012;7(9):e45151.
Crossref - Rahaie S, Emam-Djomeh Z, Razavi SH, Mazaheri M. Evaluation of aflatoxin decontaminating by two strains of Saccharomyces cerevisiae and Lactobacillus rhamnosus strain GG in pistachio nuts. Int J Food Sci Technol. 2012;47(8):1647-1653.
Crossref - Gratz S, Mykkanen H, Ouwehand AC, Juvonen R, Salminen S, El-Nezami H. Intestinal Mucus Alters the Ability of Probiotic Bacteria To Bind Aflatoxin B1 In Vitro. Appl Environ Microbiol. 2004;70(10):6306-6308.
Crossref - Eaton DL, Gallagher EP. Mechanisms of Aflatoxin Carcinogenesis. Ann Rev Pharmacol Toxicol. 1994;34(1):135-172.
Crossref - Liu Y, Galani-Yamdeu JH, Gong YY, Orfila C. A review of postharvest approaches to reduce fungal and mycotoxin contamination of foods. Compr Rev Food Sci Food Saf. 2020;19(4):1521-1560.
Crossref - Liu L, Xie M, Wei D. Biological Detoxification of Mycotoxins: Current Status and Future Advances. Int J Mol Sci. 2022;23(3):1064.
Crossref - Ranjha MMAN, Shafique B, Batool M, et al. Nutritional and Health Potential of Probiotics: A Review. Appl Sci. 2021;11(23):11204.
Crossref - Shetty PH, Jespersen L. Saccharomyces cerevisiae and lactic acid bacteria as potential mycotoxin decontaminating agents. Trends Food Sci Technol. 2006;17(2):48-55.
Crossref - Liew WPP, Nurul-Adilah Z, Than LTL, Mohd-Redzwan S. The Binding Efficiency and Interaction of Lactobacillus casei Shirota Toward Aflatoxin B1. Front Microbiol. 2018;9:1503.
Crossref - Assaf JC, Atoui A, Khoury A, Chokr A, Louka N. A comparative study of procedures for binding of aflatoxin M1 to Lactobacillus rhamnosus GG. Braz J Microbiol. 2018;49(1):120-127.
Crossref - Gul LB, Con AH, Gul O. Storage stability and sourdough acidification kinetic of freeze- dried Lactobacillus curvatus N19 under optimised cryoprotectant formulation. Cryobiology. 2020;96:122-129.
Crossref - Singh SP, Sundar K, Shetty PH. Aflatoxin [B. sub. 1] binding by microflora isolated from fermented foods. J Pure Appl Microbiol. 2016;10(1):625-631.
- Muscarella M, Iammarino M, Nardiello D, et al. Validation of a confirmatory analytical method for the determination of aflatoxins B1, B2, G1and G2 in foods and feed materials by HPLC with on-line photochemical derivatisation and fluorescence detection. Food Addi Contam: Part A Chem Anal Control Expo Risk Assess. 2009;26(10):1402-1410.
Crossref - Scheer R. Bergey’s Manual of Systematic Bacteriology. 1. Aufl. (engl.) Vol. 1: JGHolt und N. R. Krieg (Hrsg.), Seite 1-964; Vol. 2: J. G. Holt, P. H. A. Sneath, N. S. Mair, M. E. Sharpe (Hrsg.), Seite 965-1599; Williams & Wilkins, Baltimore 1984 (Vol. 1: US $87.95) und 1986 (Vol. 2: US $71.95). Pharmazie in UnsererZeit. 1990;19(4):166-166.
Crossref - Anandharaj M, Sivasankari B, Santhanakaruppu R, Manimaran M, Rani RP, Sivakumar S. Determining the probiotic potential of cholesterol-reducing Lactobacillus and Weissella strains isolated from gherkins (fermented cucumber) and south Indian fermented koozh. Res Microbiol. 2015;166(5):428-439.
Crossref - Anandharaj M, Sivasankari B. Isolation of potential probiotic Lactobacillus oris HMI68 from mother’s milk with cholesterol-reducing property. J Biosci Bioeng. 2014;118(2):153-159.
Crossref - Feng Y, Qiao L, Liu R, Yao H, Gao C. Potential probiotic properties of lactic acid bacteria isolated from the intestinal mucosa of healthy piglets. Ann Microbiol. 2017;67(3):239-253.
Crossref - Inam M, Pan S, Elken EM, et al. Antibiotic resistance of probiotics isolated from Chinese corn stover silage. J Appl Anim Res. 2023;51(1):102-114.
Crossref - Oslan SNH, Halim M, Ramle NA, et al. Improved stability of live attenuated vaccine gdhA derivative Pasteurellamul tocida B:2 by freeze drying method for use as animal vaccine. Cryobiology. 2017;79:1-8.
Crossref - Haskard CA, El-Nezami HS, Kankaanpaa PE, Salminen S, Ahokas JT. Surface Binding of Aflatoxin B1 by Lactic Acid Bacteria. Appl Environ Microbiol. 2001;67(7):3086-3091.
Crossref - Sezer C, Guven A, Bylge Oral N, Vatansever L. Detoxification of aflatoxin B1 by bacteriocins and bacteriocinogenic lactic acid bacteria. Turkish J Vet Anim Sci. 2013;37:594-601.
Crossref - Pizzolitto RP, Bueno DJ, Armando MR, Cavaglieri L, Dalcero AM, Salvano MA. Binding of Aflatoxin B1 to Lactic Acid Bacteria and Saccharomyces cerevisiae in vitro: A Useful Model to Determine the Most Efficient Microorganism. InTech eBooks. 2011.
Crossref - Marrez DA, Shahy EM, El-Sayed HS, Sultan YY. Detoxification of Aflatoxin B1 in Milk Using Lactic Acid Bacteria. J Biol Sci. 2018;18(3):144-151.
Crossref - Kumara SS, Bashisht A, Venkateswaran G, Hariprasad P, Gayathri D. Characterization of Novel Lactobacillus fermentum from Curd Samples of Indigenous Cows from Malnad Region, Karnataka, for their Aflatoxin B1 Binding and Probiotic Properties. Probiotics Antimicrob Proteins. 2018;11(4):1100-1109.
Crossref - Moller CO de A, Freire L, Rosim RE, et al. Effect of Lactic Acid Bacteria Strains on the Growth and Aflatoxin Production Potential of Aspergillus parasiticus, and Their Ability to Bind Aflatoxin B1, Ochratoxin A, and Zearalenone in vitro. Front Microbiol. 2021;12:55386.
Crossref - Nazef L, Belguesmia Y, Tani A, Prevost H, Drider D. Identification of Lactic Acid Bacteria from Poultry Feces: Evidence on Anti-Campylobacter and Anti-Listeria Activities. Poultry Science. 2008;87(2):329-334.
Crossref - Andreev N, Ronteltap M, Boincean B, Lens PNL. Lactic acid fermentation of human excreta for agricultural application. J Environ Manag. 2018;206:890-900.
Crossref - Han H, Ogata Y, Yamamoto Y, Nagao S, Nishino N. Identification of lactic acid bacteria in the rumen and feces of dairy cows fed total mixed ration silage to assess the survival of silage bacteria in the gut. J Dairy Sci. 2014;97(9):5754-5762.
Crossref - Kumari V. B. C, Huligere SS, Shbeer AM, et al. Probiotic Potential Lacticaseibacillus casei and Limosilactobacillus fermentum Strains Isolated from Dosa Batter Inhibit a-Glucosidase and a-Amylase Enzymes. Microorganisms. 2022;10(6):1195.
Crossref - Wafula EN, Muhonja CN, Kuja JO, et al. Lactic Acid Bacteria from African Fermented Cereal-Based Products: Potential Biological Control Agents for Mycotoxins in Kenya. Ren Z, ed. J Toxicol. 2022;1-17.
Crossref - Badji T, Durand N, Bendali F, et al. In vitro detoxification of aflatoxin B1 and ochratoxin A by lactic acid bacteria isolated from Algerian fermented foods. Biological Control. 2023;179:105181.
Crossref - Liu A, Zheng Y, Liu L, et al. Decontamination of Aflatoxins by Lactic Acid Bacteria. Curr Microbiol. 2020;77(12):3821-3830.
Crossref - Hoover DG, Gray RJ. Function of cell wall teichoic acid in thermally injured Staphylococcus aureus. J Bacteriol. 1977;131(2):477-485.
Crossref - Latorre-Moratalla ML, Bover-Cid S, Talon R, et al. Distribution of aminogenic activity among potential autochthonous starter cultures for dry fermented sausages. J Food Prot.2010;73(3):524-528
Crossref - El-Nezami H, Kankaanpaa P, Salminen S, Ahokas J. Physicochemical Alterations Enhance the Ability of Dairy Strains of Lactic Acid Bacteria To Remove Aflatoxin from Contaminated Media. J Food Prot. 1998;61(4):466-468.
Crossref - Banwo K, Adesina T, Aribisala O, Falade TDO. Detoxification of Aflatoxins in Fermented Cereal Gruel (Ogi) by Probiotic Lactic Acid Bacteria and Yeasts with Differences in Amino Acid Profiles. Toxins. 2023;15(3):210.
Crossref - Peltonen K, El-Nezami H, Haskard C, Ahokas J, Salminen S. Aflatoxin B1 Binding by Dairy Strains of Lactic Acid Bacteria and Bifidobacteria. J Dairy Sci. 2001;84(10):2152-2156.
Crossref - Ondiek W, Wang Y, Sun L, et al. Removal of aflatoxin b1 and t-2 toxin by bacteria isolated from commercially available probiotic dairy foods. Food Sci Technol Int. 2021;28(1):15-25.
Crossref - Ramo S, Kahala M, Joutsjoki V. Aflatoxin B1 Binding by Lactic Acid Bacteria in Protein-Rich Plant Material Fermentation. Appl Sci. 2022;12(24):12769.
Crossref - Oluwafemi F, Da-Silva FA. Removal of aflatoxins by viable and heat-killed Lactobacillus species isolated from fermented maise. J Appl Biosci. 2009;16(1):871-76.
- Marlida Y, Nurmiati N, Husmaini H, Huda N, Anggraini L, Ardani LR. The potential of lactic acid bacteria isolated from ikanbudu (fermented fish) to inhibit the growth of pathogenic fungi and detoxify aflatoxin B1. Veterinary World. 2023;16(7):1373- 1379.
Crossref - Asurmendi P, Gerbaldo G, Pascual L, Barberis L. Lactic acid bacteria with promising AFB1 binding properties as an alternative strategy to mitigate contamination on brewers’ grains. J Environ Sci Health Part B. 2020;55(11):1002- 1008.
Crossref
© The Author(s) 2023. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.