ISSN: 0973-7510
E-ISSN: 2581-690X
Escherichia coli clones, designated as extraintestinal pathogenic E. coli (ExPEC), are responsible for extraintestinal infections. Phylogenetic analysis has shown that E. coli is composed of four main phylogenetic groups (A, B1, B2 and D) and six subgroups (i.e. A0, A1, B22, B23, D1 and D2). Group A and B1 are generally associated with commensals, whereas group B2,D is associated with extra-intestinal pathotypes. In the present study, a total of 53 E. coli isolates, isolated from human clinical samples, were used. Phylogenetic grouping was done based on the PCR method using primers targeted at three genetic markers, chuA, yjaA and TspE4.C2. According to PCR-based phylotyping, subgroup B23 contained the majority of the collected isolates (40 isolates, 75.47%), followed by subgroups A1 and B22 (4 isolates for each subgroup, 7.55%), followed by subgroups D2( 3 isolates, 5.66%) and A0, D1 (1 isolate for each subgroup, 1.88%). No isolates were found to belong to group B1. Based on the results, the majority of isolates were extra-intestinal pathotypes. Therefore, the role of E. coli in human infections including urinary tract infections, septicemia, vaginitis and meningitis should be considered for further research.
Escherichia coli, Clinical Samples, Iraq.
Extraintestinal pathogenic E. coli (Ex-PEC) represents a distinct group of pathogenic E. coli that causes most of the extraintestinal E.coli infections (urinary tract infection, prostatitis, bacteremia, septicemia, and neonatal meningitis, vaginosis) human infections (Donnenberg et al., 2002; Gordon and Cowling, 2003; Al-Khaqani et al.,2016; Abdulla et al.,2016). ExPEC isolates are genetically distinct from commensal E. coli found in the intestinal flora. Phylogenetic analyses have revealed that E. coli isolates are composed of four main phylogenetic groups (A, B1, B2, and D) (Gordon, 2004 ; Al-Khaqani et al., 2016). Isolates of each of the four groups have different phenotypic features, causing their ability to exploit different sugars, antibiotic-resistance profiles and growth rate-temperature relationships . The distribution (presence/absence) of a variety of genes thought to enable a strain to cause extra-intestinal disease also varies among isolates of the four phylo-groups (Johnson et al., 2001). Several studies have shown the relation between phylogeny and pathogenicity of E.coli isolates ( Bashir et al., 2012; Escobar-Paramo et al., 2004). Bearing in mind that most commensal isolates belong to A and B1 groups (Duriez et al., 2001), Phylogenetically and epidemiologically ExPEC are potentially different from those of intestinal pathogenic and commensal isolates (Smith et al., 2007). Most of the ExPEC isolates phylogenetically belong to B2 and to a lesser extent D groups and are equipped with various virulence factors that help these isolates during different mode of infection mechanisms like adhesion, invasion of host tissues, escape host defence mechanisms, signaling and production of different toxins interfering host cellular functions thereby promoting extraintestinal infection in both normal and immune compromised hosts (Dobrindt and Hacker, 2008; Wiles et al., 2008; Al-Dahmoshi et al., 2016)
The aim of this study was to investigate the phylo-genetic groups of E. coli isolated from different clinical samples which includes urine, vaginal , seminal fluid and wound swap in Hilla, Iraq using a molecular primer.
A total of 200 various clinical samples represent by 50 urine samples from patients with urinary tract infection, 50 high vaginal swap from pregnant and non-pregnant women suffering from vaginosis, 50 seminal fluid samples of male suffering from bacteriospermia and 50 wound swabs samples. All samples were obtained from patients or individuals who were admitted to Babylon Hospital for Maternal and Pediatrics, and to Al-Hilla Surgical Teaching Hospital in Babylon city (Iraq) during the period from May to August 2016. In order to isolate E. coli, samples were directly inoculated on MacConkey agar (Himedia/India) plates. After overnight incubation at 37°C, lactose fermenting colonies, Gram-negative, oxidase negative bacilli transferred to UTI Chromogenic medium (Condalab/Spain) and Eosin methylene blue agar (Himedia/India) to confirm E. coli isolates.
DNA extraction for gram negative bacteria
Typical E. coli colonies (with metallic green color on Eosin methylene blue agar and pink color on UTI Chromogenic medium ) were grown in LB broth(Condalab/Spain) at 37ºC for 18h, and following the protocols of Favor Prep Genomic DNA Mini Kit (Blood/Cultured Cell) (Favorgen/Taiwan). The extracted DNA checked using Agarose gel electrophoresis (0.7% in TBE buffer) (Condalab/Spain) and then visualized using and gel documentation (Vilber/France).
Detection of phylogeny groups by PCR
PCR was conducted to determine the phylogenetic grouping of the isolates by targeting three genes, chuA, yjaA and TspE4.C2 using 20ìL reaction mix (IntronBio/Korea) (Clermont et al., 2000). Thermal cycler conditions were as follows: 95°C for 4 min, 30 cycles of (denaturation at 94ºC for 30sec.), (annealing at 59 ºC for 30sec.), (extension at 72ºC for 30sec). and final extension at 72ºC for 5min. Agarose gel electrophoresis (1.5% in TBE buffer) and gel documentation (Vilber/France) were used to visualized and document the PCR products. The amplicon sizes were 279 bp for chuA, 211 bp for yjaA and 152 bp for TspE4C2 were recorder using 100bp ladder (IntronBio/Korea). After electrophoresis the gel was photographed under UV light. The results allowed the classification of isolates into either one of the four major phylogroups (A, B1, B2, or D) (Abdallah et al., 2011; Gordon et al., .2008).
This study was carried out to expose the phylogeny of E. coli isolated from different clinical samples to investigate the source of these isolates whether they are intestinal or extraintestinal. However, the phylogenetic groups of E. coli isolated from clinical samples were detected by identifying the presence of specific PCR amplified fragments (chuA, yjaA, and TspE4.C2) (Figures 1,2,3) respectively.
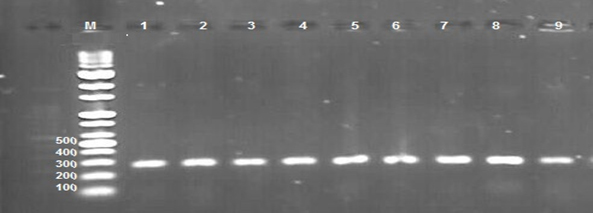
Fig. 1. 1.5% Agarose gel electrophoresis at 72 volt for 60 minutes of PCR to chuA amplicon (279bp); lane M represent DNA marker size(100bp). Lane(1-9) represent some of positive E.coli isolates.
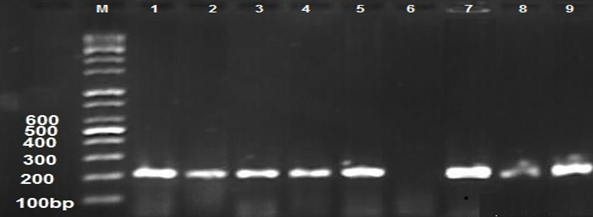
Fig. 2. 1.5% Agarose gel electrophoresis at 72 volt for 60 minutes of PCR to yja A amplicon (211bp); lane M represent DNA marker size(100bp). Lane(1-9) represent some of positive E.coli isolates.
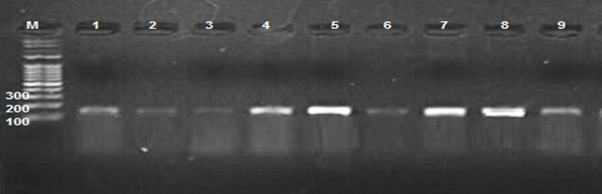
Fig. 3. 1.5% Agarose gel electrophoresis at 72 volt for 60 minutes of PCR to TspE4.C2 amplicon (152bp); lane M represent DNA marker size(100bp). Lane(1-9) represent some of posetive E.coli isolates
A total of 53 E. coli isolates represented by ( 12 vaginal ,17 UTI, 10 seminal fluid and 14 wound swap) samples and according to the presence or absence of these genes were assigned to one of four phylogenetic groups (i.e. A,B1, B2 and D) group A and B1 (intestinal groups); B2 and D (extraintestinal groups) and six subgroups (i.e. A0, A1, B22, B23, D1 and D2). According to PCR-based phylotyping the result showed that all vaginal samples, 14 urine samples, 8 of wound samples and 6 of seminal fluid samples were belong to B23 subgroup, 1 of urine samples and 3 of wound samples were belong to B22 subgroup, 2 of urine samples and 1 of seminal fluid samples were belong to D2 subgroups, 2 of wound samples and 2 of seminal fluid samples were belong to A1 subgroup, 1 of wound swap samples was belong to A0 subgroup and 1 of seminal fluid samples was belong to D1 subgroup. Also the results showed that the subgroup B23 contained the majority of the collected isolates ( 40 isolates, 75.47 %), followed by subgroups A1, B22 (4 isolates for each subgroup, 7.55 %) then subgroup D2 (3 isolates, 5.66% ) and A0, D1 (1 isolate for each subgroup, 1.88 %). No isolates were found to belong to group B1 (Table 1and 2)
Table (1):
Percentage for isolation of extraintestinal E. coli.
Sample |
Urine |
Vaginal Swab |
Seminal fluid |
Wound Swab |
Total |
|---|---|---|---|---|---|
No. of Samples |
50 |
50 |
50 |
50 |
200 |
E. coli No.(%) |
17 (34%) |
12 (24%) |
10 (20%) |
14 (28%) |
53 (26.5%) |
ExPEC, No.(%) |
17 (34%) |
12 (24%) |
8 (16%) |
11 (22%) |
48 (24%) |
Table (2):
Percentage of E. coli isolates for each phylogenetic subgroups.
| Phylogenetic groups N (%) | Phylogenetic subgroups | N (%) | Total (%) | |
|---|---|---|---|---|
| Intestinal Groups | Group A | Subgroup A0 chuA – / yjaA – / TspE4.C2 – | 1 (1.88) | 5 (9.43) |
| Subgroup A1 chuA – / yjaA + / TspE4.C2 – | 4 (7.55) | |||
| Group B1 | chuA – / yjaA – / TspE4.C2 + | 0 (0) | ||
| Extraintestinal Groups | Group B2 | Subgroup B22 chuA + / yjaA + / TspE4.C2 – | 4 (7.55) | & 48 (90.57) |
| Subgroup B23 chuA + / yjaA + / TspE4.C2 + | 40 (75.47) | |||
| Group D | Subgroup D1 chuA + / yjaA – / TspE4.C2 – | 1 (1.88) | ||
| Subgroup D2
chuA + / yjaA – / TspE4.C2 + |
3 (5.66) | |||
| Total (%) | 53 (100) | |||
The niche of commensal E. coli is the mucous layer of the colon. However, there are E. coli clones that are distinct from the intestinal commensal E. coli, possessing specific fitness and virulence attributes which allow adaptation to other niches (e.g. urinary tract, central nervous system, blood) and confer enhanced ability to cause a broad spectrum of disease in extraintestinal sites (Johnson and Russo,2005) . ExPEC isolates usually belong to phylogenetic group B2 and to a lesser extent to group D, whilst commensal isolates are derived from groups A and B1 (Escobar-Páramo et al., 2004 ). Therefore, the rapid PCR-based phylogenetic typing developed by Clermont et al.( Clermont et al .,2000) has proven useful for rapidly screening ExPEC. According to this method, about more than two-thirds of the analyzed isolates in this study belonged to group B2 (83.01%) and this is larger than the studies of Obata-Yasuoka et al, 2002 (76%) ,Watt et al,2003 (68%) and Hilbert et al, 2008 (62%). A Compared to the data reported by these researchers, group D isolates had a lower prevalence in this study (7.54% vs. 16%, 16%, and22%, respectively) , but the commensal phylogenetic group A exhibited nearest prevalence among the isolates investigated by this study (9.43% vs. 8%, 12% and 8% respectively). These differences in distribution of the phylogenetic groups among the isolates of geographically distinct populations in different studies may be due to the health status of the host, geographic climatic conditions, dietary factors, the use of antibiotics, or host genetic factors. (Duriez et al., 2001) . Based on the results of this study , the majority of isolates were extraintestinal pathotypes. Therefore, the role of E. coli in human infections including urinary tract infections, septicemia, vaginitis and meningitis should be considered for further researches.
- Abdallah KS, Cao Y, Wei DJ. Epidemiologic investigation of extraintestinal pathogenic E. coli (ExPEC) based on PCR phylogenetic group and fimH single nucleotide polymorphisms (SNPs) in China. Int J Mol Epidemiol Genet, 2011; 2: 339-353.
- Abdulla, A.A., Al-Dahmoshi, H.O.M., Abed, T.A. and Muttaleb, W.H. Characterization of multidrug resistant carbapenemases-producing Escherichia coli and Klebsiella pneumoniae isolates from urinary tract infection. Journal of Chemical and Pharmaceutical Sciences, 2016; 9(3):1116-1120.
- Al-Dahmoshi, H.O.M., Almamoori, A.M.J. and Al-Khafaji, N.S.K. Investigation of CusCFBA pump among uropathogenic Escherichia coli (UPEC) Isolated from women with cystitis, Iraq. Asian Journal of Pharmaceutical and clinical research, 2016; 10(1).
- Al-Khaqani, M.M., Alwash, M.S. and Al-Dahmoshi, H.O. Phylogrouping, antibiotics susceptibility and biofilm formation among cervico-vaginal Escherichia coli (CVEC) isolated for female in Hilla City, Iraq. Malaysian Journal of Microbiology, 2016; 12(4).
- Bashir S, Haque A, Sarwar Y, Anwar A, Anwar M. Virulence profile of different phylogenetic groups of locally isolated community acquired uropathogenic E. coli from Faisalabad region of Pakistan. Ann Clin Microbiol Antimicrob. 2012; 11:23.
- Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol., 2000; 66: 4555-4558.
- Dobrindt U. and Hacker J. Targeting virulence traits: potential strategies to combat extraintestinal pathogenic E. coli infections. Current opinion in microbiology, 2008; 11: 409-413.
- Donnenberg M. Escherichia coli virulence mechanisms of versatile pathogen. Elsevier Science, San Diego Calif. : Academic Press; 2002. pp. xxi–xxv.
- Duriez P, Clermont O, Bonacorsi S, Bingen E, Chaventre A, Elion J, Picard B, Denamur E. Commensal Escherichia coli isolates are phylogenetically distributed among geographically distinct human populations. Microbiology, 2001; 147: 1671-1676.
- Escobar-Paramo P, Clermont O, Blanc-Potard A, Bui H, Le Bouguenec C, Denamur E. A specific genetic background is required for acquisition and expression of virulence factors in Escherichia coli. Mol Biol Evol., 2004; 21: 1085-1094.
- Gordon D, Cowling A. The distribution and genetic structure of Escherichia coli in Australian vertebrates: host and geographic effects. Microbiology, 2003; 149: 3575-3586
- Gordon DM, Clermont O, Tolley and H, Denamur E. Assigning Escherichia coli isolates to phylogenetic groups: multi-locus sequence typing versus the PCR triplex method. Environ Microbiol, 2008; 10: 2484-2496.
- Gordon DM. The influence of ecological factors on the distribution and genetic structure of Escherichia coli. In Escherichia coli and Salmonella typhimurium. American Society for Microbiology 2004; [http:// www.ecosal.org/ecosal/index.jsp].
- Hilbert DW, Paulish TE, Mordechai E, Adelson ME, Trama JP. O serogroups, phylogeny, and virulence factors of cervicovaginal and rectal Escherichia coli isolates Eur. J Clin Microbiol Infect Dis, 2008; 27(12):1265-8.
- Johnson JR, Delavari P, Kuskowski M, Stell AL. Phylogenetic distribution of extraintestinal virulence-associated traits in Escherichia coli. J Infect Dis, 2001; 183:78-88.
- Johnson JR, Russo TA. Molecular epidemiology of extraintestinal pathogenic (uropathogenic) Escherichia coli. Int J Med Microbiol., 2005; ; 295(6-7):383-404
- Obata-Yasuoka M, Ba-Thein W, Tsukamoto T, Yoshikawa H, Hayashi H. Vaginal Escherichia coli share common virulence factor profiles, serotypes and phylogenywith other extraintestinal E. coli. Microbiology,, 2002; 148(Pt 9):2745-52.
- Orskov F, Orskov I. Escherichia coli serotyping and disease in man and animals. Can J Microbiol, 1992; 38: 699-704.
- Smith J.L., Fratamico P.M. and Gunther N.W. Extraintestinal pathogenic Escherichia coli. Foodborne pathogens and disease, 2007; 4: 134-163.
- Watt S, Lanotte P, Mereghetti L, Moulin-Schouleur M, Picard B, Quentin R. Escherichia coli isolates from pregnant women and neonates: intraspecies genetic distribution and prevalence of virulence factors. J Clin Microbiol., 2003; 41(5):1929-35.
- Wiles T.J., Kulesus R.R. and Mulvey M.A. Origins and virulence mechanisms of uropathogenic Escherichia coli. Experimental and molecular pathology, 2008; 85: 11-19.
© The Author(s) 2017. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.


