ISSN: 0973-7510
E-ISSN: 2581-690X
Vancomycin-resistant Enterococci(VRE) were common among Enterococcus. faecalis and Enterococcus faecium. Teicoplanin resistance or sensitivity can determine the VRE phenotypes whether VanA (VanR/TecR) or VanB (VanR/TecS). Linezolid resistance among VRE regards an newly emerged health problem. Infection with LRVRE or TRLRVRE pushan hazardous alert for hard to heal illness. Twenty eight Enterococcus spp. isolates were recovered from children diarrhea after their inoculation on m-EI chromogenic agar. Antibiotics susceptibility and phenotypic detection of antibiotics resistance were performed according to CLSI 2016. The results revealed 92.86% resistance to rifampin, 85.71% to erythromycin. VRE were 46.42%, TRE were 25% and LRE were 35.71% while co-existed resistance for Vancomycin/Teicoplanin/Linezolid(TRLRVRE) were detected 25% in. concern antibiotics resistant patterns, the MDR compile (85.7%) while XDR compile (10.7%) and there is no PDR among Enterococcus. spp. isolates were PDR. The presentstudy conclude that VanA and VanB phenotypes were common among MDR and XDR and although there is no using of linezolid but the emergence of TRLRVRE isolates were stated.
Enterococcus spp., VRE, MDR, XDR, LRVRE, TRLRVRE.
Enterococci werea Gram-positive, non-spore-forming,catalase-negativefacultative-anaerobe, Which normally dwellthe alimentary tract of humans. even if Enterococcus spp. is a coexistence organism of the intestinal tract. However although, It may be the causative agent of diarrhea in the elderly and children and immune compromised patients1,2. Enterococci, particularly relevant Enterococcus faecium and Enterococcus faecalis, have arise as objects of importance because of the distinctness of resistant strains of many drugs3-5. Enterococcus which includes some of nosocomial multidrug-resistant organisms. Vancomycin-resistant enterococci (VRE) is now one of the leading causes of nosocomial infections and represent approximately one-third of Enterococcus isolates6-8. There are three main patterns of resistance: Multi-drug resistance (MDR) was indicated as acquired non-susceptibility to asminimumas one agent in three or more antimicrobial classes, extensive-drug resistance (XDR) was clear as non-susceptibility to at least one agent in all but two or fewer antimicrobial categories and pan-drug resistance (PDR) was defined as resistance to all classesof anti-microbial9-12. Vancomycin and teicoplanin resistance via one or more of nine genes (vanA-vanE, vanG, vanL, vanM and vanN) which express for enzymes needed for the synthesis of new peptidoglycan precursors and enzymes that disrupt the normal d-Ala-d-Ala-ending precursors13-15. Enterococci resistant to erythromycin by main two mechanism: enzyme production like ribosomal methylases (coded for by erm genes) methylate the bacterial ribosome, impairing the binding of macrolide and macrolide efflux, coded for by mef genes 16]. Eflux pumps encoded by tetKand tetLwere responsible for tetracyclines17. Three mechanisms were described well includes mutation in gyrA gene, production of NorA efflux pump and encoding for Qnr proteins which guard DNA-gyrase by diminishing DNA binding of the quinolone and succeeding formation of the quinolone–gyrase complex. Tell yet Rifampicin-resistance get up from a range of mutations in the rpoB gene that encodes for the polymerase RNA b-subunit. Two mechanism of resistance were well described among Enterococcus sp. to linezolid : genes in which mutations occurencoding the 23S rRNA, (which is an important part of the drug-binding site on the ribosome) and Enzymatic modification of the 23S rRNA by methylase17-24. In this study aims to check the antibiotic resistance patterns along with resistance phenotypes of diarrheal Enterococcus faecalis and Enterococcus faecium.
Sample Collection and Processing
Fifty eight stool samples (diarrhea) were collected from children with diarrhea with age ranged from 1-7 years. Swab were used to take the sample and put it in brain heart infusion broth for transportation and incubated at 37°C for 24 hrs. and then inoculated to mEI chromogenic agar25.
Cultivation on m-EI chromogenic agar
Agar chromogenic mEI is a chromogenetic agar for recovery and distinctionof faecalis and faecium enterococci. It containnutrients and cycloheximidefor fungi inhibition. Incubation for 18-24 hours and then Enterococcus faecium itGrowth will appears greenish-blue, while give blue colonies for Enterococcus faecalis.
Antibiogram
Antibiotic susceptibility test were done according to CLSI 201626 using standard disk diffusion method upon Muller-Hinton agar after normalization of broth to 0.5 McFarland (1×108 CFU/ml at OD=0.08).
Biosafty Aspects
The biosafty aspects include decontamination of swabs, broth, contaminated disposable and culture medium27.
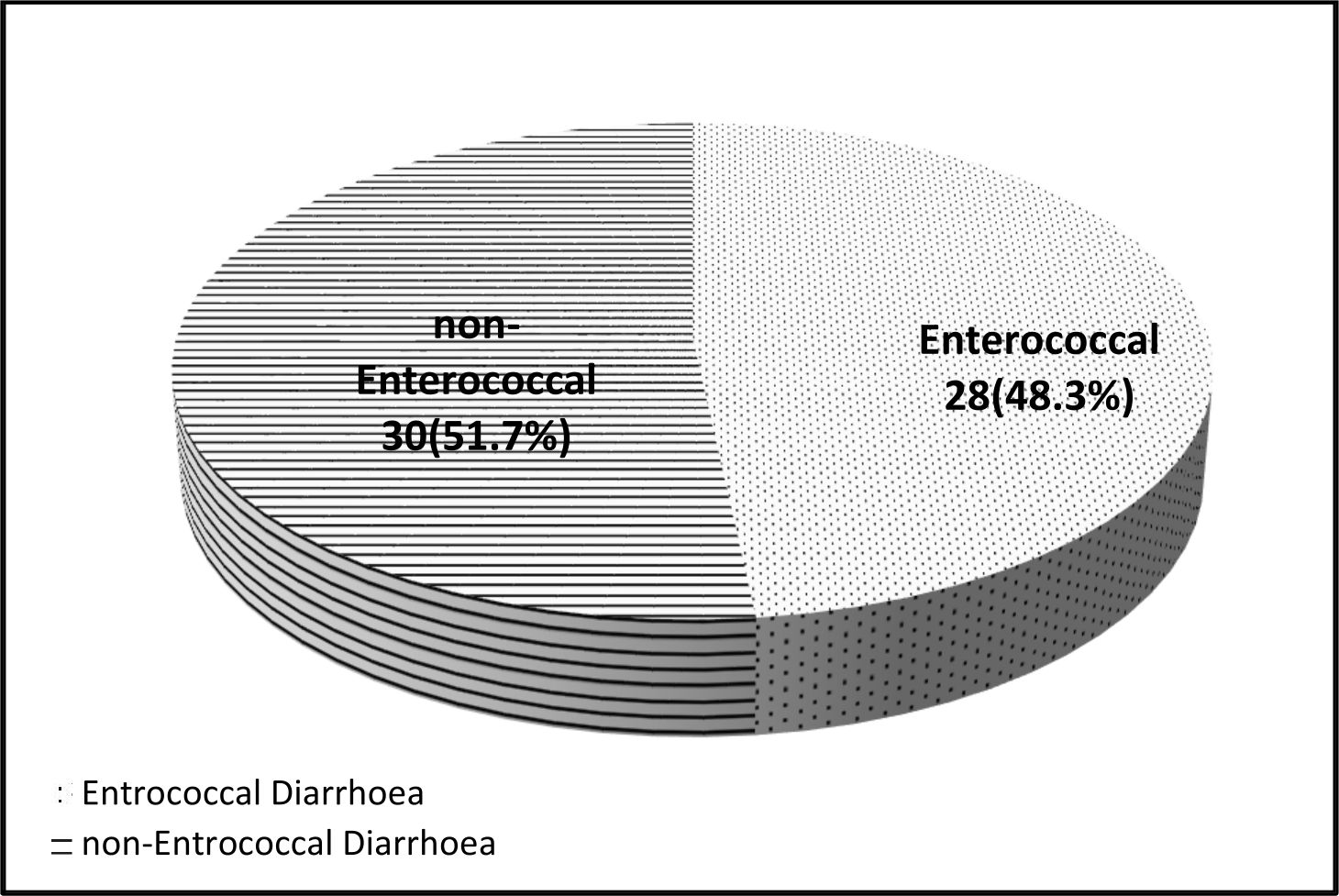
The result of Enterococcus sp. Isolation revealed that, enterococcal diarrhea compile 28(48.3%) Fig. 1. Enterococcus spp. is intestinal opportunistic bacterium with virulence possibilities like protease gelatinase (GelE). It is not naturally virulent but their resistance arrays of antibiotics classes leading to infections in susceptible individual like immuno-compromised, children and elderly and cancer patients. Infected patients with diarrhea can be the source of MDR-enterococci especially VRE28,29.
Enterococcus spp. regards reservoir of intrinsic and inherently resistance to various antibiotics classes. The results of the current study publicized different resistance percentage as rifampin (92.86%), erythromycin (85.71%), nitrofurantion (64.29%), ciprofloxacin (60.71%), tetracyclin (57.14%), penicillin (53.57%), vancomycin (46.43%), linezolid (35.71%), teicoplanin (25%) and doxycyclin (17.86%) table (1).
Table (1):
Antibiotics resistance percentage among Enterococcus spp.
Antibiotic |
Symbol |
Potency (μg) |
Resistance % |
|---|---|---|---|
Rifampin |
RA |
5 |
92.86 |
Erythromycin |
E |
15 |
85.71 |
Nitrofurantion |
F |
300 |
64.29 |
Ciprofloxacin |
CIP |
5 |
60.71 |
Tetracycline |
TE |
30 |
57.14 |
Penicillin |
P |
10 |
53.57 |
Vancomycin |
VA |
30 |
46.43 |
Linezolid |
LNE |
30 |
35.71 |
Teicoplanin |
TEC |
30 |
25 |
Chloramphenicol |
CHL |
30 |
21.34 |
Doxycycline |
DO |
30 |
17.86 |
Table (1) show the results for antibiotic resistance among Enterococcus spp. Our results in accordance with many Iraqi studies like Khalid (2016)30, Chabuck et al., (2011)31, Al-Marjani (2013)32 and Al-Halaby AH, Al-Hashimy (2016)33 who found (72-100%) of enterococci resistance to rifampin respectively. Many studies around the world also stated similar results, resistance to enterococci were (76-100%)34-36. The most common mechanism of resistance to rifampin is mutation in b subunit of RNA polymerase (encodes by rpoB)37. Resistance to erythromycin were (85.71%) and it is quite same those stated in another studies30-33,38-40. Co-existence of triple resistance to vancomycin/teicoplanin/linezolid were present in 7/13 (53.84%) of vancomycin resistant enterococci (VRE) table (2). Our results is the first who stated co-existence of resistance to vancomycin/teicoplanin/linezolid in Iraq while many studies in Iraq and neighboring countries not state such resistance30-40. The most common phenotypes of vancomycin resistance among VRE are VanA and VanB which related to vanA and vanB genotypes. Van A characterized by their co-resistance to both vancomycin and teicoplanin while VanB confer only resistance to vancomycin41. Our results stated both phenotypes, VanA in 7/13 (53.84%) while VanB in 6/13 (46.16%).
Resistance of enterococci to linezolid (LRE) were very rare and single cases documented around the world. Also the Co-existed resistance to vancomycin and linezolid ( LRVRE) and vancomycin, teicoplanin and linezolid (TRLRVRE) were note documented yet in Iraq and this study seem the first to report TRLRVRE phenotypically. The results revealed that 7/28 (25%) of enterococci were TRLRVRE or LRVRE table (2). Linezolid resistance may be appear after treatment with linezolid while many cases reported the resistance in patients without prior use of linezolid42-46.
Table (2):
Co-existence resistance among Enterococcus spp.
Antibiotics co-existence |
No.( %) |
VancomycinR total (VRE) |
13/28 (46.42) |
TeicoplaninR total (TRE) |
7/28 (25.00) |
LinezolidR total (LRE) |
10/28 (35.71) |
VancomycinR/TeicoplaninS/ LinezolidS |
6/28 (24.00) |
VancomycinR/TeicoplaninR/ LinezolidS (TRVRE) |
0/28 (0.00) |
VancomycinR/TeicoplaninR/LinezolidR (TRLRVRE) |
7/28 (25.00) |
VancomycinS/TeicoplaninR/ LinezolidR (TRLRE) |
0/28 (0.00) |
VancomycinS/TeicoplaninS/ LinezolidR |
3/28 (10.71) |
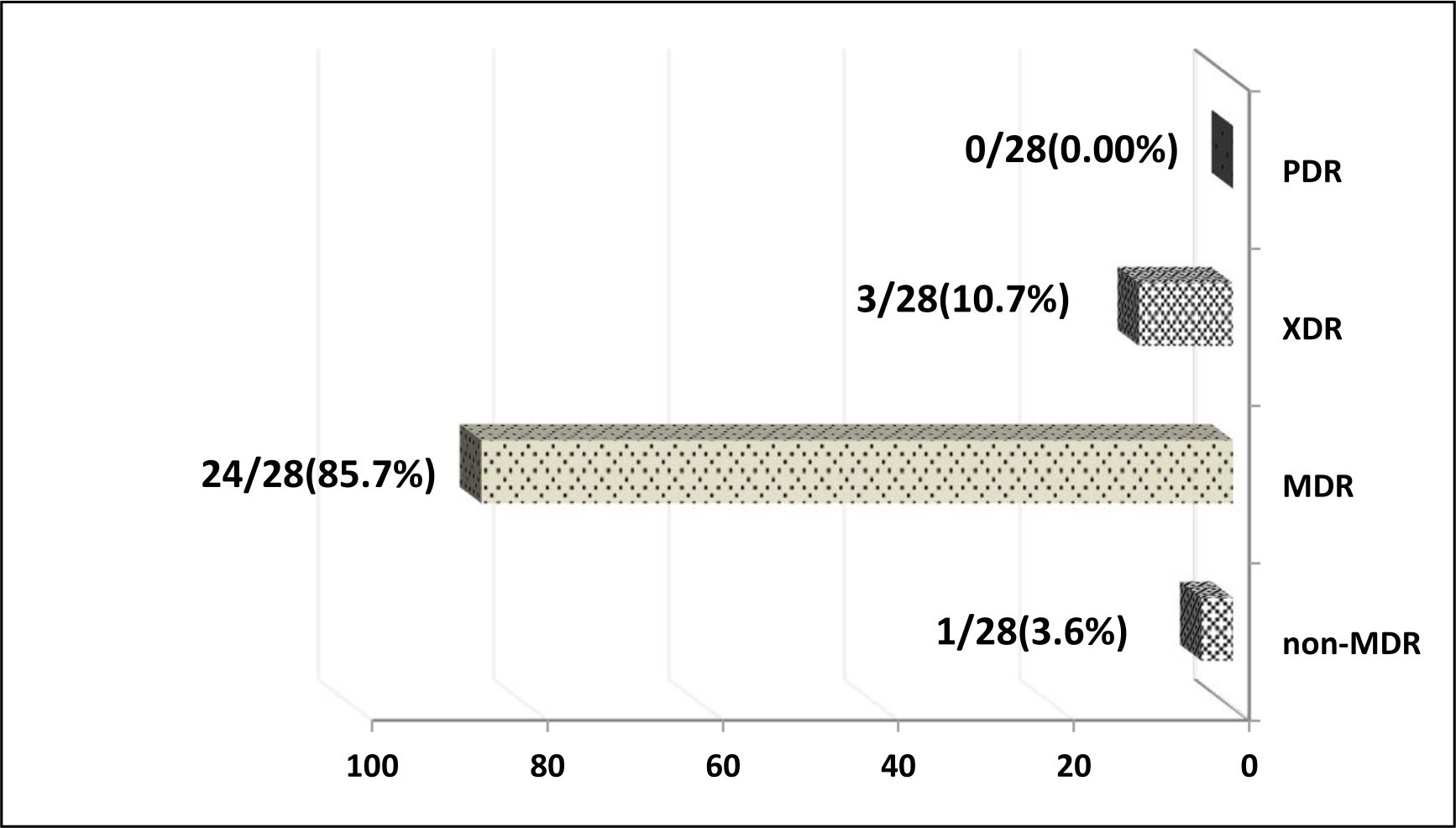
Concern the resistance patterns, MDR, XDR and PDR, the results revealed that 1/28 (3.6%), 24/28(85.7%), 3/28(10.7%) and 0/28 (0.00%) of enterococci isolates were non-MDR, MDR, XDR and PDR respectively.
Different percentage of MDR-enterococci were stated in many studies (28-63%)47-50. XDR-enterococci were also stated in many studies and compile ( 8-35%) of isolated enterococci. The resulted multidrug or extensive drug resistance, due to many factors such as antibiotic pressures or antibiotics abuse, can leads to costly, hard to cure, prolonged illness and high mortality infections51,52.
The current study conclude that VanA and VanB phenotypes were common among MDR and XDR and although there is no using of linezolidbut the emergence of TRLRVRE isolates were stated.
The authors declare that there are no conflicts of interest.
- Sah R, Khadka S, Shah DS, Adhikari M, Shrestha N, Kattel HP, Sharma S, Mishra SK, Parajuli K, Sherchand JB, Shah NP. Vanconycin resistant Enterococcus faecalis causing diaeehea in renal transplant patient. International Educational Applied Scientific Research Journal, 2017, 18; 2(9).
- Fisher K, Phillips C. The ecology, epidemiology and virulence of Enterococcus. Microbiology, 2009, 1; 155(6):1749-57.
- K hn I, Iversen A, Burman LG, Olsson-Liljequist B, Franklin A, Finn M, Aarestrup F, Seyfarth AM, Blanch AR, Taylor H, Caplin J. Epidemiology and ecology of enterococci, with special reference to antibiotic resistant strains, in animals, humans and the environment: example of an ongoing project within the European research programme. International Journal of Antimicrobial Agents, 2000; 14(4):337-42.
- Mundy LM, Sahm DF, Gilmore M. Relationships between enterococcal virulence and antimicrobial resistance. Clinical microbiology reviews, 2000; 13(4):513-22.
- Miller WR, Murray BE, Arias CA. Emergence and management of drug-resistant enterococcal infections. Future Medicine Ltd.
- Arias CA, Murray BE. The rise of the Enterococcus: beyond vancomycin resistance. Nature Reviews Microbiology, 2012; 10(4):266.
- Hidron AI, Edwards JR, Patel J, Horan TC, Sievert DM, Pollock DA, Fridkin SK. Anti-microbial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006–2007. Infection Control & Hospital Epidemiology, 2008; 29(11):996-1011.
- Sievert DM, Ricks P, Edwards JR, Schneider A, Patel J, Srinivasan A, Kallen A, Limbago B, Fridkin S. Antimicrobial-resistant pathogens associated with healthcare-associated infections summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009–2010. Infection Control & Hospital Epidemiology, 2013; 34(1):1-4.
- Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson Liljequist B, Paterson DL. Multidrug resistant, extensively drug resistant and pandrug resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clinical microbiology and infection, 2012; 18(3):268-8.
- Sifaoui F, Arthur M, Rice L, Gutmann L. Role of penicillin-binding protein 5 in expression of ampicillin resistance and peptidoglycan structure in Enterococcus faecium. Antimicrobial agents and chemotherapy, 2001; 45(9):2594-7.
- Ono S, Muratani T, Matsumoto T. Mechanisms of resistance to imipenem and ampicillin in Enterococcus faecalis. Antimicrobial agents and chemotherapy, 2005; 49(7):2954-8.
- Sarti M, Campanile F, Sabia C, Santagati M, Gargiulo R, Stefani S. Polyclonal diffusion of beta-lactamase-producing Enterococcus faecium. Journal of clinical microbiology, 2012; 50(1):169-72.
- San Millan A, Depardieu F, Godreuil S, Courvalin P. VanB-type Enterococcus faecium clinical isolate successively inducibly resistant to, dependent on, and constitutively resistant to vancomycin. Antimicrobial agents and chemotherapy, 2009; 53(5):1974-82.
- Xu X, Lin D, Yan G, Ye X, Wu S, Guo Y, Zhu D, Hu F, Zhang Y, Wang F, Jacoby GA. vanM, a new glycopeptide resistance gene cluster found in Enterococcus faecium. Anti-microbial agents and chemotherapy, 2010; 54(11):4643-7.
- Lebreton F, Depardieu F, Bourdon N, Fines-Guyon M, Berger P, Camiade S, Leclercq R, Courvalin P, Cattoir V. D-Ala-D-SerVanN-type transferable vancomycin resistance in Enterococcus faecium. Antimicrobial agents and chemotherapy, 2011; 55(10):4606-12.
- Leclercq R. Mechanisms of resistance to macrolides and lincosamides: nature of the resistance elements and their clinical implications. Clinical Infectious Diseases, 2002; 34(4):482-92.
- Chopra I, Roberts M. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiology and molecular biology reviews, 2001; 65(2):232-60.
- Werner G, Fleige C, Ewert B, Laverde-Gomez JA, Klare I, Witte W. High-level ciprofloxacin resistance among hospital-adapted Enterococcus faecium (CC17). International journal of antimicrobial agents, 2010; 35(2):119-25.
- Lףpez M, Tenorio C, Del Campo R, Zarazaga M, Torres C. Characterization of the mechanisms of fluoroquinolone resistance in vancomycin-resistant enter-ococci of different origins. Journal of Chemotherapy, 2011; 23(2):87-91.
- Yasufuku T, Shigemura K, Shirakawa T, Matsumoto M, Nakano Y, Tanaka K, Arakawa S, Kawabata M, Fujisawa M. Mechanisms of and risk factors for fluoroquinolone resistance in clinical Enterococcus faecalis isolates from patients with urinary tract infections. Journal of clinical microbiology, 2011; 49(11):3912-6.
- Kristich CJ, Little JL. Mutations in the b subunit of RNA polymerase alter intrinsic cephalosporin resistance in Enterococci. Antimicrobial agents and chemotherapy, 2012; 56(4):2022-7.
- Boumghar-Bourtchaן L, Dhalluin A, Malbruny B, Galopin S, Leclercq R. Influence of recombination on development of mutational resistance to linezolid in Enterococcus faecalis JH2-2. Antimicrobial agents and chemotherapy, 2009; 53(9):4007-9.
- Diaz L, Kiratisin P, Mendes RE, Panesso D, Singh KV, Arias CA. Transferable plasmid-mediated resistance to linezolid due to cfr in a human clinical isolate of Enterococcus faecalis. Antimicrobial agents and chemotherapy, 2012; 56(7):3917-22.
- Bi R, Qin T, Fan W, Ma P, Gu B. The emerging problem of linezolid-resistant enterococci. Journal of global antimicrobial resistance, 2018; 13:11-9.
- U.S. Environmental Protection Agency. Method 1600: Enterococci in water by membrane filtration using membrane enterococcus indoxyl –D- glucoside agar (mEl]. Publication EPA-821- R-02-022. USEPA Office of Water, Office of Science and Technology, USEPA, Washington, DC, 2002.
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing. 26th ed. CLSI supplement M100S. Wayne, PA: Clinical and Laboratory Standards Institute; 2016.
- Panel BB. Guidelines for safe work practices in human and animal medical diagnostic laboratories. Morbidity and Mortality Weekly Report. 2012; 6:61.
- Pinholt M, ״stergaard C, Arpi M, Bruun NE, Schרnheyder HC, Gradel KO, Sרgaard M, Knudsen JD. Incidence, clinical characteristics and 30-day mortality of enterococcal bacteraemia in Denmark 2006–2009: a population-based cohort study. Clinical Microbiology and Infection, 2014; 20(2):145-51.
- Sah R, Khadka S, Shah DS, Adhikari M, Shrestha N, Kattel HP, Sharma S, Mishra SK, Parajuli K, Sherchand JB, Shah NP. VANCOMYCIN RESISTANT ENTEROCOCCUS FAECALIS CAUSING DIARRHEA IN RENAL TRANSPLANT PATIENT. International Educational Applied Scientific Research Journal. 2017; 2(9).
- Ubeda C., Taur Y., Jeng R. R., Equinda M. J., Son T., Samstein M., et al. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. The Journal of Clinical Investigation, 2010; 120(12):4332–4341.
- Khalid HM. Molecular Detection of Virulence Factors of Enterococcus Faecalis Isolated From Urine Samples in Duhok City, Kurdistan Region/Iraq. Science Journal of University of Zakho, 2016, 11;4(1):63-72.
- Chabuck ZA, Al-Charrakh AH, Al-Sa’adi MA. Prevalence of vancomycin resistant enterococci in Hilla City, Iraq. Med. J. Babylon–8 (3). 2011.
- AL-Marjani MF. vanA in vancomycin-resistant Enterococcus faecalis isolated in Baghdad. African Journal of Microbiology Research, 2013; 7(2):115-9.
- Al-Halaby AH, Al-Hashimy AB. Assessment of Biofilm Production and Antibiotic Pattern in E. faecium and E. faecalis isolated from Some UTI Iraqi Patients. Int. J. Curr. Microbiol. App. Sci., 2016; 5(11):161-72.
- Shehabi AA, Badran EF, Alnasra NA. The emergence of antimicrobial resistance in enterococci isolates from infants: A review study. The International Arabic Journal of Antimicrobial Agents, 2015, 12;4(3).
- Jahansepas A, Aghazadeh M, Rezaee MA, Hasani A, Sharifi Y, Aghazadeh T, Mardaneh J. Occurrence of Enterococcus faecalis and Enterococcus faecium in Various Clinical Infections: Detection of Their Drug Resistance and Virulence Determinants. Microbial Drug Resistance, 2018; 24(1):76-82.
- Du X, Hua X, Qu T, Jiang Y, Zhou Z, Yu Y. Molecular characterization of Rifr mutations in Enterococcus faecalis and Enterococcus faecium. Journal of Chemotherapy. 2014; 26(4):217-21.
- XiaotingHua, TingtingQu, Xi Li, Qiong Chen, ZhiRuan, Yunsong Yu. Global Effect of rpoB Mutation on Protein Expression in Enterococcus faecium. Jundishapur Journal of Microbiology, 2016; 9:12.
- Sharifi Y, Hasani A, Ghotaslou R, Naghili B, Aghazadeh M, Milani M, Bazmany A. Virulence and antimicrobial resistance in enterococci isolated from urinary tract infections. Advanced pharmaceutical bulletin, 2013; 3(1):197.
- Yadegarynia D, Roodsari SR, Arab-Mazar Z. Evaluation of Antimicrobial Susceptibility Among Enterococcus Species by E-Test Method at Khatam-ol-Anbia Hospital During 2013–2014. Archives of Clinical Infectious Diseases, 2016; 11(1).
- Heidari H, Hasanpour S, Ebrahim-Saraie HS, Motamedifar M. High Incidence of Virulence Factors Among Clinical Enterococcus faecalis Isolates in Southwestern Iran. Infection & chemotherapy. 2017; 49(1):51-6.
- Qu TT, Zhang JL, Zhou ZH, Wei ZQ, Yu YS, Chen YG, Li LJ. Heteroresistance to teicoplanin in Enterococcus faecium harboring the vanA gene. Journal of clinical microbiology. 2009; 47(12):4194-6.
- Kumar S, Bandyoapdhyay M, Chatterjee M, Mukhopadhyay P, Poddar S, Banerjee P. The first linezolid-resistant Enterococcus faecium in India: High level resistance in a patient with no previous antibiotic exposure. Avicenna journal of medicine, 2014; 4(1):13.
- de Almeida LM, de Araתjo MR, Iwasaki MF, Sacramento AG, Rocha D, da Silva LP, Pavez M, de Brito AC, Ito LC, Gales AC, Lincopan N. Linezolid resistance in vancomycin-resistant Enterococcus faecalis and Enterococcus faecium isolates in a Brazilian hospital. Antimicrobial agents and chemotherapy, 2014; 58(5):2993-4.
- Klare I, Fleige C, Geringer U, Th rmer A, Bender J, Mutters NT, Mischnik A, Werner G. Increased frequency of linezolid resistance among clinical Enterococcus faecium isolates from German hospital patients. Journal of global antimicrobial resistance, 2015; 3(2):128-31.
- Niebel M, Perera M, Shah T, Marudanayagam R, Martin K, Oppenheim BA, David MD. Emergence of linezolid resistance in hepatobiliary infections caused by Enterococcus faecium. Liver Transplantation, 2016; 22(2):201-8.
- Gupta S. Emergence of linezolid resistance in clinical isolates of vancomycin-resistant enterococci. International Journal of Advanced Medical and Health Research, 2016; 3(2):107.
- Bibalan MH, Eshaghi M, Sadeghi J, Asadian M, Narimani T, Talebi M. Clonal diversity in Multi Drug Resistant (MDR) Enterococci isolated from fecal normal flora. International journal of molecular and cellular medicine, 2015; 4(4):240.
- Daniel DS, Lee SM, Dykes GA, Rahman S. Public health risks of multiple-drug-resistant Enterococcus spp. in Southeast Asia. Applied and environmental microbiology. 2015; 81(18):6090-7.
- Bhatt P, Patel A, Sahni AK, Praharaj AK, Grover N, Chaudhari CN, Das NK, Kulkarni M. Emergence of multidrug resistant enterococci at a tertiary care centre. Medical Journal Armed Forces India, 2015; 71(2):139-44.
- Basak S, Singh P, Rajurkar M. Multidrug resistant and extensively drug resistant bacteria: A study. Journal of pathogens. 2016;2016.
- Carlson AL, Pruetpongpun N, Buppajarntham A, Apisarnthanarak A. Controlling nosocomial transmission of drug-resistant pathogens at different endemic stages in a resource-limited setting. Infection Control & Hospital Epidemiology. 2016; 37(9):1114-6.
© The Author(s) 2019. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.