ISSN: 0973-7510
E-ISSN: 2581-690X
Staphylococcus aureus is the microbe implicated in the causation of numerous nosocomial and community-acquired ailments. Physicians have been facing enormous challenges day by day while treating methicillin-resistant Staphylococcus aureus (MRSA) efficiently because it shows extensive resistance to wide number of antibiotics. MRSA is frequently treated using antibiotics like macrolide-lincosamide-streptogramin B. Clindamycin is a useful alternative drug. Nevertheless, inducible resistance to clindamycin cannot be detected by standard in vitro sensitivity tests. The aim of the study was to identify MRSA isolates from clinical samples from a tertiary care centre in North India that possessed both constitutive as well as inducible clindamycin resistance. This is a one year (July 2023-June 2024) cross-sectional study carried out in the Department of Microbiology of Maharaja Agrasen Medical College, Agroha (Hisar). 1554 pus samples were collected, processed and antibiotic susceptibility was assessed using disk diffusion method. Cefoxitin disk was utilised for identification of the isolates as MRSA and by employing D-test, inducible clindamycin resistance was identified. S. aureus was isolated in 234 samples, of which 182 (78%) were MRSA and 52 (22%) were MSSA. Among MRSA strains, 36.2% showed inducible clindamycin resistance, 6.6% showed constitutive phenotype and 31.9% showed MS phenotype. It should be emphasized that it is possible to successfully cure MRSA infections using clindamycin but only after the simple and affordable D-test is employed in a bid to rule out inducible clindamycin resistance. Therefore, to avert unsuccessful therapy in MRSA infections, D-test should be carried out on a regular basis.
Constitutive Phenotype cMLSB, D-test, Inducible Phenotype iMLSB, MRSA, MS Phenotype
Worldwide, Staphylococcus aureus is believed to be one of the main culprits of nosocomial and community-acquired infections.1 It is the most prevalent isolate linked to skin diseases and soft tissue infections.2 If appropriate treatment is not given promptly, then, sepsis, amputation and disfigurement can result from skin infections as well as soft tissue infections. The length of hospital stay for the patient also gets extended.3 There has been a significant increase in resistance among Staphylococcus against various antimicrobial agents.4 Methicillin resistance is one such mechanism. Globally, over the past ten years, methicillin-resistant Staphylococcus aureus (MRSA) has shown an increasing upsurge in its prevalence.5
It is difficult to cure methicillin-resistant Staphylococcus aureus because of its pervasive resistance to different antibiotics. Antibiotics named macrolide-lincosamide-streptogramin B (MLSB) are most frequently utilized for addressing MRSA, clindamycin being commonly preferred.6 This is because clindamycin has good pharmacokinetic properties, available as both oral and parenteral formulations, inhibits toxin production, relatively cheap and it has excellent soft tissue permeability. With the passage of time and overuse of antimicrobials, S. aureus has additionally started developing resistance to MLSB antibiotics too.
Resistance towards clindamycin in S. aureus can be seen in any of the two ways – induced or constitutive.7 The most commonly observed mechanism of this kind of resistance is modification of its target site. Erythromycin ribosome methylase (erm) genes are particularly responsible for this modification. The expression of erm gene can occur constitutively (cMLSB), meaning that the strains are resistant to both clindamycin and erythromycin, or inducibly (iMLSB), meaning that the strains exhibit clindamycin sensitivity but erythromycin resistance.8 The msrA gene mediates another pathway of resistance (MS phenotype) leading to efflux of antibiotic. Drug inactivation (encoded by lnu genes) also contributes to development of resistance.9 Thus, three different phenotypes are shown by S. aureus strains for MLSB antibiotics.10
This cross-sectional study was undertaken in the Department of Microbiology at Maharaja Agrasen Medical College, Agroha (Hisar), Haryana, over a time frame of one year from July 2023-June 2024. 234 S. aureus isolates were obtained from 1554 pus samples evaluated for culture and sensitivity. Pus samples that were collected in the microbiology laboratory from individuals of all ages and genders who were suspected of having infections involving skin and soft tissues, whether inpatients or outpatients were included in the study. The study did not include S. aureus isolates from samples other than pus. The study was begun after receiving ethical approval from institutional ethics committee. After being entered and assembled in an MS Excel spreadsheet, the data was transferred to SPSS (Statistical Package for Social Sciences) version 30.0 for additional assessment.
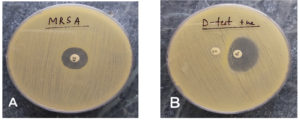
Standard culture media such as Blood agar and MacConkey agar were put to use to culture each sample, and they were kept under incubation at 37 °C for about 24 to 48 hours. First of all, the isolates obtained were recognized as S. aureus with the help of standard laboratory protocols such as growth on culture media (colony morphology), gram staining and various biochemical reactions (such as catalase test-positive, slide coagulase test and tube coagulase test-positive).11 Once the isolates were confirmed to be S. aureus, the antimicrobial susceptibility testing (AST) was performed on Muller Hinton agar (MHA) (HiMedia Pvt. Ltd., India) by utilising Kirby Bauer’s disk diffusion method. Interpretation of the results was made on the basis of CLSI (Clinical and Laboratory Standards Institute).12 The antibiotics included for evaluation were – ampicillin (10 µg), erythromycin (15 µg), amikacin (30 µg), cefoxitin (30 µg), ceftriaxone (30 µg), gentamicin (10 µg), co-trimoxazole (sulphamethoxazole/trimethoprim) (23.75/1.25 µg), linezolid (30 µg), penicillin-G (10 µg), tetracycline (30 µg), oxacillin (1 µg), norfloxacin (10 µg), ciprofloxacin (5 µg), vancomycin and clindamycin (2 µg). Detection of MRSA among the isolates was done with the aid of cefoxitin disk (30 µg). Phenotypic validation of the isolates as MRSA was done only if they showed a zone of inhibition of 21 mm or less surrounding the cefoxitin disk (Figure 1A). The strain utilized as the control was S. aureus ATCC 25923.
Figure 1. (A). MRSA. Cefoxitin disk showing <21 mm zone of inhibition. (B). Clindamycin disk-surrounded by a zone of inhibition resembling shape of D that flattens in the direction of erythromycin disk
In accordance with CLSI standards (2024), the S. aureus isolates demonstrating erythromycin resistance were next subjected to the D-test to determine whether they possessed constitutive or inducible resistance to clindamycin. Muller Hinton agar plates were lawn cultured with S. aureus 0.5 McFarland standard bacterial suspension in order to perform the D-test. Next, the clindamycin disk (2 µg) and the erythromycin disk (15 µg) were set aside 15 mm from one another (edge to edge). After that an incubation of 37 °C was given to the plates and kept in an aerobic environment for 18 to 24 hours.12
The following interpretations were made of three distinct S. aureus phenotypes-
- Constitutive Phenotype (cMLSB)- Isolates showing clindamycin (zone size <14 mm) and erythromycin (zone size ≤13 mm) resistance in addition to development of a circular shaped inhibition zone encircling clindamycin disk, if existent.
- Inducible Phenotype (iMLSB)- Isolates with erythromycin resistance (zone size <13 mm) and displaying clindamycin sensitivity (zone size ≥21 mm), where the clindamycin disk forms a D-shaped inhibition zone that flattens out close to the erythromycin disk (Figure 1B).
- MS Phenotype- Erythromycin resistant (zone size ≤13 mm) isolates displaying clindamycin drug sensitivity (zone size ≥21 mm) and demonstrating a clindamycin disk encircled by a circular zone of inhibition.
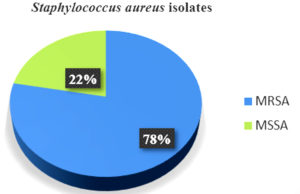
Of 234 S. aureus isolates, 182 (78%) were MRSA and 52 (22%) were MSSA (Figure 2). The isolates’ sensitivity to erythromycin along with the other antibiotics in the panel was assessed by disk diffusion test. Of the total isolates, 160 (68.3%) displayed erythromycin resistance.
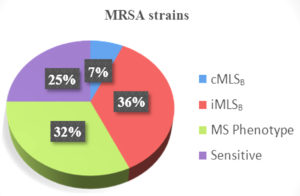
D-test was conducted on the isolates that demonstrated erythromycin resistance. Among MRSA, 36.2% of the isolates exhibited inducible resistance towards clindamycin with a positive result for D-test, constitutive phenotype was displayed by 6.6% solates (cMLSB) and 31.9% isolates showed MS Phenotype (Figure 3). The percentage of MSSA isolates with constitutive and induced clindamycin resistance was 7.7% and 23%, respectively. MS phenotype was shown by 15.4% MSSA isolates. There were distinct variations in constitutive and inducible clindamycin resistance while comparing MRSA strains with MSSA strains (Table).
Table:
Comparative evaluation of cMLSB, iMLSB and MS Phenotypes within MRSA and MSSA strains
Phenotypes |
MRSA |
MSSA |
Total |
|---|---|---|---|
E-R, C-R (cMLSB) |
12 (6.6%) |
4 (7.7%) |
16 (6.8%) |
E-R, C-S, D+ (iMLSB) |
66 (36.2%) |
12 (23%) |
78 (33.3%) |
E-R, C-S, D- (MS Phenotype) |
58 (31.9%) |
8 (15.4%) |
66 (28.2%) |
E-S, C-S |
46 (25.2%) |
28 (53.8%) |
74 (31.6%) |
Total |
182 (100%) |
52 (100%) |
234 (100%) |
MSSA = Methicillin-sensitive Staphylococcus aureus, MRSA = Methicillin-resistant Staphylococcus aureus, C = Clindamycin, E = Erythromycin, S = Sensitive, R = Resistant, D+ = D-test positive, D- = D-test negative, iMLSB = Inducible Phenotype, cMLSB = Constitutive Phenotype and MS Phenotype
Since S. aureus is typically prevalent on the skin’s surface and in environment, it is frequently present in pus samples and wound swabs. Effective treatment of S. aureus is hampered by the emergence of resistance for various antibiotics in the bacteria. Therefore, it often becomes essential to ascertain the antibiotic susceptibility of a clinical isolate, so that the infected patients receive the best possible antimicrobial therapy. MRSA and other severe staphylococcal infections have been treated with clindamycin. Nevertheless, staphylococcal isolates with inducible phenotypes are susceptible to developing clindamycin resistance. It has been noted that these isolates spontaneously develop constitutively resistant mutants when clindamycin is administered either in vitro or in vivo.13
We observed a high percentage of MRSA (78%) in the course of our study as compared to MSSA (22%) among 234 S. aureus isolates. The results were comparable to those discovered by Mama et al., Bala et al. and Godebo et al.2,3,14 Our study yielded higher outcomes as compared to other research studies.1 This could be due to the inclusion of pus samples only. Of the total 234 isolates, 160 (68.3%) showed resistance to erythromycin. The D-test was applied in an attempt to look for inducible clindamycin resistance (ICR) for every single isolate that showed erythromycin resistance. Among MRSA, 66 (36.2%) isolates showcased inducible clindamycin resistance, as indicated by a D-test being positive (iMLSB phenotype). A negative D-test result was found in 58 (31.9%) isolates (MS phenotype). 12 isolates (6.6%) exhibited the cMLSB trait, or constitutive resistance. Similar outcomes have been demonstrated by various other studies such as those by Prabhu et al., Deotale et al., Thapa et al., Ghosh et al., where inducible clindamycin resistance among MRSA was reported as 37.5%, 45%, 36.5% and 41.3%, respectively.4,15-17 However, higher percentage (52.3%) was reported by Seifi et al.18 Additionally, we discovered that MRSA had a greater percentage of iMLSB phenotypes (36.2%) than cMLSB phenotypes (6.6%). In our study, among MSSA isolates, constitutive and inducible resistance was displayed by 4 (7.7%) and 12 (23%) isolates. 8 (15.4%) isolates displayed MS phenotype.
Furthermore, MRSA has higher value of inducible clindamycin resistance and MS phenotypes (36.2% and 31.9% respectively) than MSSA (23% and 15.4% respectively). The results are in consensus with researches conducted by Yilmaz et al, Deotale et al, Gadepalli et al, where the percentage of inducible resistance was 24.4%, 27.6%, and 30% in MRSA and 14.4%, 1.6% and 10% in MSSA respectively.13,15,19 Ajantha et al. in their study have shown that MRSA have considerably higher prevalence of inducible resistance.20 Conversely, a handful of researches have revealed that MSSA exhibits a greater percentage of inducible resistance than MRSA.21,22
Clindamycin is a great choice for treatment on outpatient basis owing to its high absorption when consumed orally. Nevertheless, ICR is typically missed by standard in vitro clindamycin susceptibility testing. Thus, therapy with clindamycin in these cases will not be effective. For patients to be managed effectively, ICR must be identified otherwise therapy with clindamycin can lead to treatment failure in such cases.23 A simple approach for determining inducible clindamycin resistance is the double disk diffusion test, often known as the D-test. ICR is illustrated by D-shaped inhibition zone surrounding clindamycin along with the presence of flattening near erythromycin.24 Therefore, inclusion of D-test in routine workflow of laboratory will enable us to steer physicians on appropriate usage of clindamycin for dermatological diseases and soft tissue infections.
The overwhelming majority of nosocomial and community-acquired infections involving soft tissues and skin are triggered by MRSA. The inability to effectively combat MRSA is because they have started developing resistance to many antibiotics. Amongst the MLSB family, clindamycin is a preferable option for the management of MRSA due to its enhanced tissue penetration, reduced cost and fewer adverse effects. The iMLSB strains can progressively evolve yielding mutants that depict constitutive resistance when the patient is given clindamycin for therapy, both in vitro and in vivo. Henceforth, in an effort to avoid therapy failure, it is imperative that S. aureus isolates resistant to erythromycin undergo continuous surveillance for clindamycin sensitivity using the D-test.
ACKNOWLEDGMENTS
The authors would like to thank the Management of Maharaja Agrasen Medical College, Agroha (Hisar), Haryana, for their support.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
Both authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This study was approved by the Institutional Ethics Committee, Maharaja Agrasen Medical College, Agroha (Hisar), Haryana, with reference number MAMC/IEC/2024/22.
- Goudarzi M, Kobayashi N, Dadashi M, et al. Prevalence, genetic diversity, and temporary shifts of inducible clindamycin resistance Staphylococcus aureus Clones in Tehran, Iran: a molecular- epidemiological analysis from 2013 to 2018. Front Microbiol. 2020;11:663.
Crossref - Mama M, Aklilu A, Misgna K, Tadesse M, Alemayehu E. Methicillin- and Inducible Clindamycin-Resistant Staphylococcus aureus among Patients with Wound Infection Attending Arba Minch Hospital, South Ethiopia. Int J Microbiol. 2019;2019(1):2965490.
Crossref - Bala R, Kaur N, Gupta N, et al. Detection of Inducible Resistance to Clindamycin among Methicillin Resistant and Sensitive strains of Staphylococcus aureus from India. J Pure Appl Microbiol. 2021;15(4):1957-1962.
Crossref - Prabhu K, Rao S, Rao V. Inducuble clindamycin resistance in Staphylococcal aureus isolated from clinical samples. J Lab Physicians. 2011;3(1):25-27.
Crossref - Boucher HW, Corey GR. Epidemiology of Methicillin- resistant Staphylococcus aureus. Clin Infect Dis. 2008;46(Suppl 5):S344-349.
Crossref - Delialioglu N, Aslan G, Ozturk C, Baki V, Sen S, Emekdas G. Inducible clindamycin resistance in staphylococci isolated from clinical samples. Jpn J Infect Dis. 2005;58(2):104-106.
Crossref - Lim HS, Lee H, Roh KH, Yum JH, Yong D, Lee K. Prevalence of inducible clindamycin resistance in staphylococcal isolates at Korean tertiary care hospital. Yonsei Med J. 2006;47(4):480-484.
Crossref - Gupta V, Datta P, Rani H, Chander J. Inducible clindamycin resistance in Staphylococcus aureus: a study from North India. J Postgrad Med. 2009;55(3):176-179.
Crossref - Leclercq R. Mechanism of resistance to macrolides and lincosamides: naure of resistance elements and their clinical implications. Clin Infect Dis. 2002;34(4):482-492.
Crossref - Lata S, Bala R, Jindal N, Gupta N. In Vitro Study of Constitutive and Inducible Clindamycin Resistance in Staphylococcus aureus with Reference to Methicillin Resistant Staphylococcus aureus: Experience from Tertiary Care Hospital in Punjab. Ind J Pub Health Res Dev. 2020;11(2):314-317.
Crossref - Gary WP, Church DL, Hall GS. Koneman’s Color Atlas and Textbook of Diagnostic Microbiology: 7th Edition, Washington C. 2017.
- CSLI M100. Performance standards for Antimicrobial Susceptibility Testing. M100, 34th ed. Clinical and Laboratory Standards Institute. 2024: 216.
- Yilmaz G, Aydin K, Iskender S, Caylan R, Koksal I. Detection and prevalence of inducible clindamycin resistance in staphylococci. J Med Microbiol. 2007;56(3):342-5.
Crossref - Godebo G, Kibru G, Tassew H. Multidrug-resistant bacterial isolates in infected wounds at Jimma University Specialized Hospital, Ethiopia. Ann Clin Microbiol Antimicrob. 2013;12:17.
Crossref - Deotale V, Mendiratta DK, Raut U, Narang P. Inducible clindamycin resistance in Staphylococcus aureus isolated from clinical samples. Ind J Med Microbiol. 2010;28(2):124-126.
Crossref - Thapa D, Pyakurel S, Thapa S, et al. Staphylococcus aureus with inducible clindamycin resistance and methicillin resistance in a tertiary hospital in Nepal. Trop Med Health. 2021;49(1):99.
Crossref - Ghosh S, Banerjee M. Methicillin resistance & inducible clindamycin resistance in Staphylococcus aureus. Indian J Med Res. 2016;143(3):362-364.
Crossref - Seifi N, Kahani N, Askari E, Mahdipour S, Naderi NM. Inducible clindamycin resistance in Staphylococcus aureus isolates recovered from Mashhad, Iran. Iran J Microbiol. 2012;4(2):82-86.
- Gadepalli R, Dhawan B, Mohanty S, Arti Kapil, Das BK, Chaudhry R. Inducible clindamycin resistance in clinical isolates of Staphylococcus aureus. Indian J Med Res. 2006;123(4):571-573.
- Ajantha GS, Kulkarni RD, Shetty J, Shubhada C, Jain P. Phenotypic detection of inducible clindamycin resistance among Staphylococcus aureus isolates by using the lower limit of recommended inter-disk distance. Indian J Pathol Microbiol. 2008;51(3):376-378.
Crossref - Schreckenberger PC, Ilendo E, Ristow KL. Incidence of constitutive and inducible clindamycin resistance in Staphylococcus aureus and Coagulase-negative staphylococci in a community and a tertiary care hospital. J Clin Microbiol. 2004;42(6):2777-2779.
Crossref - Levin TP, Suh B, Axelrod P, Truant AL, Fekete T. Potential clindamycin resistance in clindamycin-susceptible, erythromycin-resistant Staphylococcus aureus: Report of a clinical failure. Antimicrob Agents Chemother. 2005;49(3):1222-4.
Crossref - Siberry GK, Tekle T, Carrol K, Dick J. Failure of clindamycin treatment of Methicillin-resistant Staphylococcus aureus expressing inducible clindamycin resistance in vitro. Clin Infect Dis. 2003;7(9):1257-1260.
Crossref - Saffar H, Rajabiani A, Abdollahi A, Habibi S, Baseri Z. Frequency of inducible clindamycin resistance among gram-positive cocci in a tertiary hospital, Tehran, Iran. Iran J Microbiol. 2016;8(4):243-248.
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.