ISSN: 0973-7510
E-ISSN: 2581-690X
The present study was conducted for the phenotyping of antibiotic resistance patterns among patients infected with Klebsiella pneumoniae, isolated from different clinical sites of the patients admitted to the Medical City Teaching Laboratories in Baghdad, Iraq, and to study the frequencies of the blaCTX-M, blaTEM, and blaOXA genes in the extended-spectrum b-lactamase (ESBL)-producing isolates. A total of 20 out of 35 (57.14%) K. pneumoniae isolates collected from different clinical samples were identified as ESBL producers using the combination disk test (CDT) against six types of antibiotics, as suggested by the Clinical and Laboratory Standards Institute. All K. pneumoniae isolates were observed for ESBL positivity using the CDT method and screened for blaTEM, blaCTX-M, and blaOXA genes by PCR using a specific primer. In total, 19/20 (95.0%) ESBL-positive isolates harbored the TEM genes, 18/20 (90.0%) carried CTX-M, while the blaOXA gene, for the first time in Baghdad city, was not reported in any of the isolates. A high occurrence of ESBL-producing K. pneumoniae was observed in our study based on the analysis of the TEM and CTX-M genes. Although molecular methods are more reliable in identifying ESBL production, routine clinical screening for ESBL-producing K. pneumoniae by phenotypic methods, such as CDT tests, must be introduced and encouraged in clinical settings because of its low cost.
K. pneumoniae, blaOXA, CTX-M, blaTEM, ombination disk test (CDT)
Klebsiella pneumoniae is the causative agent of opportunistic diseases in humans, such as respiratory tract infection, burn inflammation, wound inflammation, septicemia, diarrhea, and liver abscesses1. The urinary tract is commonly infected by K. pneumoniae, following Escherichia coli2. The resistance of K. pneumoniae to antibiotics, such as beta-lactam antibiotics, is mainly caused by the production of a broad-spectrum of beta-lactamases (ESBL), which is an important problem leading to increased infections in hospitals; the resistance results from an alteration of the permeability barrier or the bacterial target site represented by a penicillin-binding protein, and by the changes in the outer membrane protein3. Beta-lactam antibiotics are frequently used to treat bacterial infections. The antibiotic groups in this category, including penicillins, cephalosporins, and carbapenems, are associated with the emergence of beta-lactam-mediated bacterial resistance, which subsequently results in the development of ESBL-producing bacteria. ESBLs are enzymes that mediate resistance to an extended spectrum of antibiotics, for example, third-generation cephalosporins, and monobactams, such as aztreonam4. ESBLs can be classified into three main types, TEM, SHV, and CTX-M.5. The isolates of the Enterobacteriaceae family showing plasmid-mediated lactamase production have the blaTEM gene, which hydrolyzes penicillins, first-generation cephalosporins, as well as the blaCTX-M gene, which preferentially hydrolyzes cefotaximes6. The other ESBL gene type is SHV, which is responsible for the plasmid-mediated resistance to ampicillin. Most ESBLs are SHV or TEM enzyme derivatives most frequently present in enterobacteria, such as K. pneumoniae and E. coli7. This study aimed to isolate and identify the types ESBLs phenotypically and genotypically produced by K. pneumoniae isolated from different clinical samples (urine, blood, stool, sputum, and burn) from patients admitted to the Medical City Teaching laboratories, Baghdad, Iraq. In addition, phenotyping and molecular detection of the three beta-lactam genes (OXA, SHV, and CTX-M) were performed by studying the prevalence of various ESBL genotypic patterns in ESBL-producing bacterial isolates.
Phenotypic methods for ESBL pattern determination
Thirty-five isolates of K. pneumoniae bacteria were previously acquired and identified from different clinical samples from patients admitted to the Medical City Teaching Laboratories, Baghdad, Iraq between November 2020 and January 2021. The isolates were phenotypically investigated for ESBL production using the CDT as a screening test, per the guidelines of the Clinical and Laboratory Standards Institute (CLSI)8. Resistance to 6 types of drugs at different concentrations (Bioanalyse, Turkey) (Table 2) was tested using the disk diffusion method, following the manufacturer’s instructions9.
Table (1):
Primer design of three ESBL genes and its product yields.
| Genes | Primer Sequence (5′ to 3′) | Size of product bp | References |
|---|---|---|---|
| TEM- F | TCCTTGAGAGTTTTCGCCCC | 550 | Design in this study (D) |
| TEM- R | TGACTCCCCGTCGTGTAGAT | ||
| CTX-M- F | AAGCACGTCAATGGGACGAT |
500 |
|
| CTX-M- R | GTTGGTGGTGCCATAGCCA | ||
| OXA- F | TTGCACTTGATAGTGGTGTGA | no band | |
| OXA- R | AGTGAGTTGTCAAGCCAAAAAGT |
Genotypic methods for ESBL resistance determination
Extraction of K. pneumoniae genome
DNA extraction was performed using the steps mentioned in PrestoTM Mini gDNA Bacteria Kit (Geneaid, Taiwan). After extraction, the gel electrophoresis method was used to detect the presence of the K. pneumoniae genome. The selected genome of K. pneumoniae was then further investigated for the molecular detection of three types of ESBL genes, blaCTX-M-D1, blaTEM-D1, and bla OXA-D-1.
Polymerase chain reaction (PCR) technique for beta-lactamase encoding gene detection
PCR was performed according to the manufacturer’s instructions (Bioneer, South Korea). A PCR assay, which was designed in this study and gave the different sizes of bands, was used to detect CTX-M, TEM, and OXA genes (Table 1) (Sentebiolab, Turkey). The following PCR program was used: one cycle of 3 min, initial denaturation at 94°C, followed by 40 cycles of denaturation at 93°C for 1 min, annealing at 54°C for 1 min, extension at 72°C for 1 min, and a final extension step for 10 min at 72°C. Then, 5 ìL each of the amplified PCR products was loaded onto an agarose gel along with a standard molecular weight DNA ladder (Promega, USA) and subjected to electrophoresis as described by a previous study10.
Table (2):
Phenotyping and genotyping patterns of isolated K. pneumoniae from different clinical samples.
| No | Position | CTX | CTC | FEP | FEC | CAZ | CZC | Phenotyping patterns | Genotyping patterns | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Interpretation | TEM | CTX | OXA | |||||||||
| 1 | Urine | 22 | 20 | 27 | 26 | 11 | 16 | ESBL positive | + | + | – | |
| 2 | Urine | 35 | 21 | 30 | 29 | 29 | 31 | ESBL positive | + | + | – | |
| 3 | Urine | 0 | 12 | 18 | 22 | 11 | 22 | ESBL positive | + | + | – | |
| 4 | Urine | 17 | 11 | 26 | 22 | 0 | 11 | ESBL positive | + | + | – | |
| 5 | Urine | 31 | 30 | 32 | 20 | 22 | 27 | ESBL positive | + | + | – | |
| 6 | Urine | 0 | 15 | 17 | 21 | 17 | 23 | ESBL positive | + | + | – | |
| 7 | Urine | 0 | 0 | 9 | 7 | 0 | 16 | ESBL positive | + | + | – | |
| 8 | stool | 25 | 20 | 25 | 23 | 20 | 13 | ESBL positive | + | + | – | |
| 9 | Urine | 19 | 10 | 16 | 10 | 10 | R | ESBL positive | + | + | – | |
| 10 | Blood | 20 | 15 | 25 | 25 | 20 | 10 | ESBL positive | + | + | – | |
| 11 | Urine | 15 | R | 14 | R | R | R | ESBL positive | + | + | – | |
| 12 | Sputum | 25 | 2 | 2 | 15 | 15 | 8 | ESBL positive | + | + | – | |
| 13 | Urine | 30 | 25 | 25 | R | 1 | R | ESBL positive | + | + | – | |
| 14 | Urine | 20 | 15 | 2 | 2 | 15 | R | ESBL positive | + | + | – | |
| 15 | Burn | 25 | 15 | 25 | 20 | 15 | 7 | ESBL positive | – | – | – | |
| 16 | Blood | 20 | 15 | 20 | 15 | 14 | 15 | ESBL positive | + | – | – | |
| 17 | Urine | 5 | R | R | R | 7 | 10 | ESBL positive | + | + | – | |
| 18 | Urine | 25 | 20 | 25 | 20 | 20 | 20 | ESBL positive | + | + | – | |
| 19 | Urine | 8 | R | 7 | R | 0 | 8 | ESBL positive | + | + | – | |
| 20 | Blood | 25 | 15 | 22 | 20 | 17 | 13 | ESBL positive | + | + | – | |
| Total | N | 20/20 | 20/19 | 20/18 | 20/0 | |||||||
| % | 100% | 95% | 90% | 0.0% | ||||||||
CTX 30; cefotaxime; CTC 40; cefotaxime / clavulanic Acid; FEP 30; cefepime; FEC40; cefepimeclavulanic Acid; CAZ 30 ; ceftazidime; CZC 40 ; ceftazidime / clavulanic Acid, R;resistance, S,sensitive
Statistical analysis
Data analysis were performed using MS Excel software (version 2010).
Phenotypic analysis for antibiotic resistance determination
Determination using CDT
In total, 35 K. pneumoniae isolates were tested for ESBL production. The results showed that 20 (57.14%) isolates were ESBL positive, which was indicated by an increase in the score of the inhibition zone diameter e” 5 mm with clavulanic acid than without (Table 2), while 15 (42.85%) isolates were ESBL negative, which was indicated by a decrease in score of inhibition zone < 5 mm with clavulanic acid than without (data not shown). The results of the present study are in agreement with those of a previous study11, in which 58.8% of the isolates produced ESBLs, as determined by CDT. The results of the present study also agree with another previous study12, that showed that the resistance rate of ESBL-producing K. pneumoniae isolated from different clinical samples was 88.23%, as determined by the disk diffusion test (DDT), used as a screening test for ESBL detection. It was noted that CDT was easily performed and immediately interpreted. The specificity and sensitivity of this procedure were initially recorded to be 96% and 100%, respectively5. The high percentages of ESBL-producing isolates of K. pneumoniae detected in this study may be due to patients taking large amounts of third-generation cephalosporins, sometimes without the guidance of a physician, and the widespread abuse of antibiotic administration by patients as well as physicians.
Molecular detection of the ESBL genes
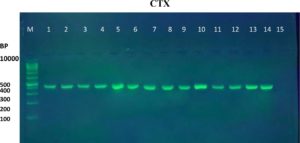
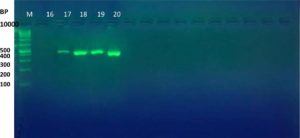
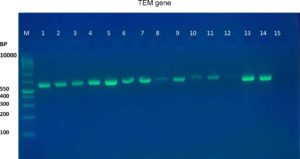
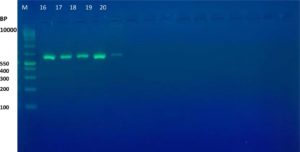
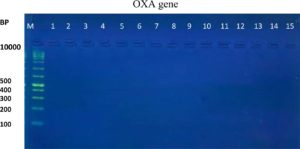
The results of the ESBL phenotyping of the investigated isolates were highly correlated with the genotyping test results. Based on the results of the CDT test, only 20 (57.14%) out of the 35 K. pneumoniae isolates were ESBL-producing. All three K. pneumoniae genes associated with ESBL production, namely the blaTEM, blaCTX-M, and blaOXA genes, were screened using PCR. Genotypically, 95% (19/20) of the ESBL-positive isolates produced the TEM ESBLs, whereas 90% (18/20) produced CTX-M ESBLs. In contrast, no isolate showed the production of OXA ESBLs (Table 2 and Figure 1-6). According to the type of the sample, the results of this study showed that genotypically, the highest number of resistance patterns were observed among the K. pneumoniae isolated from urine samples. Locally, the findings of this study were consistent with those of a study in Najaf City13 that used PCR methods and reported that 60.1% of the isolated K. pneumoniae carried CTX-M genes. In another study in Erbil City, the ESBL-producing K. pneumoniae showed a lower resistance rate than those obtained in this study, with 64.7% harboring blaTEM ESBL genes while 41.1% harboring the blaCTX-M genes14. Another study performed in Najaf City focused on studying ESBL genotypic patterns among patients with urinary tract infections without kidney disease and those with chronic kidney disease. The study revealed that the percentages of the blaTEM- and blaCTX-M-harboring ESBL-positive K. pneumoniae isolates were 83.3%, 88.2%, 61.1%, and 6.4%15. Genotypically, the percentage found in the current study was higher than that found in Turkey in a previous study12 that demonstrated that the TEM-type ESBLs were found in 73.33% of K. pneumoniae isolates from different clinical samples. In a study conducted in India, 115 (38%) of 304 K. pneumoniae isolates harbored blaTEM16. With respect to the ESBL blaOXA gene, our results showed that no isolate harbored the OXA genes. The production of beta-lactamases, which inactivate and hydrolyze b-lactam antibiotics, has become the most important resistance mechanism of different bacterial species, particularly among the Enterobacteriaceae family17. In this study, ESBL-positive isolates were detected using CDT and PCR. Twenty K. pneumoniae isolates were positive in the CDT test and 19/20 and 18/20 K. pneumoniae isolates were positive for the TEM and CTX genes, respectively, as determined by PCR. The observation of negative ESBLs in the two isolates using PCR confirmed the accuracy of the DDT. SHV and TEM b-lactamases are particularly present in K. pneumoniae and E. coli but may be found in other members of the Enterobacteriaceae family and in non-enteric microorganisms, such as Acinetobacter species18. Rapid laboratory identification of these strains is necessary because of their resistance to available antibiotics, the genes responsible for which can be passed to other strains. Molecular typing can identify the type of ESBLs that exist in each isolate. Molecular identification and detection of beta-lactamases are important for the accurate epidemiological detection of antimicrobial resistance.
Fig. 1. Gel electrophoresis of PCR product of CTX-M gene in K. pneumoniae isolates, M= DNA molecular size markers (100 bp), 1 to 15 represents number of isolates, 1% agarose gel at 5 Vol /cm for 1:15 hours
Fig. 2. Gel electrophoresis of PCR product of CTX-M gene in K. pneumoniae isolates, M= DNA molecular size markers (100 bp), 16 to 20 represents number of isolates, 1% agarose gel at 5 Vol /cm for 1:15 hours.
Fig. 3. Gel electrophoresis of PCR product TEM gene in K. pneumoniae isolates, M= DNA molecular size markers (100 bp), 1 to 15 represents number of isolates, 1% agarose gel at 5 Vol /cm for 1:15 hours
Fig. 4. Gel electrophoresis of PCR product TEM gene in K. pneumoniae isolates, M= DNA molecular size markers (100 bp), 16 to 20 represents number of isolates, 1% agarose gel at 5 Vol /cm for 1:15 hours
Fig. 5. Gel electrophoresis of PCR product OXA gene in K. pneumoniae isolates, M= DNA molecular size markers (100 bp), 1 to 15 represents number of isolates, 1% agarose gel at 5 Vol /cm for 1:15 hours
In this study, the isolated K. pneumoniae exhibited high resistance to most antibiotics, especially the beta-lactam group, and were considered multidrug-resistant bacteria. Furthermore, K. pneumoniae produced the TEM and CTX-M ESBLs at high rates of occurrence, reaching 95 % and 90%, respectively. This highlights the need to adopt strict protocols related to antibiotic administration in hospitals to control the emergence of extensive antibiotic resistance in different species of bacteria. Because of the significance of ESBL-producing K. pneumoniae and the difficulties involved in the therapy of infections caused by these bacteria, clinical laboratories should adopt a simple test based on the suggestions of the CLSI for the rapid identification and confirmation of ESBL production in Enterobacteriaceae.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
All authors declared that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
ETHICS STATEMENT
Not applicable
AVAILABILITY OF DATA
All data sets generated or analyzed during this study are included in the manuscript.
- Zhang S, Yang G, Ye Q, Wu Q, Zhang J, Huang Y. Phenotypic and genotypic characterization of Klebsiella pneumonia isolated from retail foods in China. Frontiers in microbiology, 2018;9:289.
Crossref - Li JJ, Sheng ZK, Deng M, et al. Epidemic of Klebsiella pneumonia ST11 clone coproducing KPC-2 and 16S rRNA methylase RmtB in a Chinese University Hospital. BMC infectious diseases. 2012;12:373.
Crossref - Aghamohammad S, Badmasti F, Solgi H, Aminzadeh Z, Khodabandelo Z, Shahcheraghi F. First report of extended-spectrum betalactamase-producing Klebsiella pneumonia among fecal carriage in Iran: high diversity of clonal relatedness and virulence factor profiles. Microbial Drug Resistance. 2020; 26(3):261-269.
Crossref - Wayne PA. Clinical and Laboratory Standards Institute: Performance standards for antimicrobial susceptibility testing: 20th informational supplement. CLSI document 2010; M100-S20. 2010.
- Bradford PA. Extended-spectrum b-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clinical microbiology reviews. 2001;14(4):933-951.
Crossref - Rawat D, Nair D. Extended-spectrum b-lactamases in Gram Negative Bacteria. J Glob Infect Dis. 2010; 2(3): 263-274.
Crossref - Sharma M, Pathak S, Srivastava P. Prevalence and antibiogram of Extended Spectrum b-Lactamase (ESBL) producing Gram negative bacilli and further molecular characterization of ESBL producing Escherichia coli and Klebsiella spp. Journal of Clinical Diagnostic Research. 2013;7(10):2173-2177.
Crossref - Magiorakos AP, Srinivasan A, Carey RT, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clinical microbiology and infection. 2012;18(3):268-281.
Crossref - Al Sweih N, Jamal W, Rotimi VO. Spectrum and antibiotic resistance of uropathogens isolated from hospital and community patients with urinary tract infections in two large hospitals in Kuwait. Medical Principles and Practice. 2005;14(6):401-407.
Crossref - Mishra AK, Sharma K, Misra RS. Isozyme and PCR-based genotyping of epidemic Phytophthora colocasiae associated with taro leaf blight. Archives of Phytopathology and Plant Protection. 2010;43(14):1367-1380.
Crossref - Abusaiba THH, Alshammari MMM, Kadhum HA, Yousif MG. Extended Spectrum Beta-Lactamase Producing Klebsiella pneumonia Isolated from Patients with Urinary Tract Infection in Al-Najaf Governorate-Iraq. International Journal of Advances in Science, Engineering and Technology (IJASEAT). 2020;8(1):13-16.
- Bali EB, Acik L,Sultan N. Phenotypic and molecular characterization of SHV, TEM, CTX-M and extended-spectrum beta-lactamase produced by Escherichia coli, Acinobacter baumannii and Klebsiella isolates in a Turkish hospital. Afr J Microbiol Res. 2010;18;4(8):650-654.
- Shabaa RA. Detection of CTX-M-1 gene Among Klebsiella pneumonia Isolates in An Najaf Province. Iraqi journal of biotechnology. 2014;13(2):128-133.
- Pishtiwan AH, Khadija KM. Prevalence of blaTEM, blaSHV, and blaCTX-M genes among ESBL-producing Klebsiella pneumonia and Escherichia coli isolated from thalassemia patients in Erbil, Iraq. Mediterranean journal of hematology and infectious diseases. 2019; 11(1):e2019041.
Crossref - Majeed HT, Aljanaby AA. Antibiotic susceptibility patterns and prevalence of some extended spectrum beta-lactamases genes in gram-negative bacteria isolated from patients infected with urinary tract infections in Al-Najaf City, Iraq. Avicenna journal of medical biotechnology. 2019; 11(2):192-201.
- Gautama V, Thakur A, Sharma M, et al. Molecular characterization of extended-spectrum b-lactamases among clinical isolates of Escherichia coli &Klebsiella pneumoniae: a multi-centric study from tertiary care hospitals in India. The Indian journal of medical research. 2019;149(2):208-215.
Crossref - Akcam FZ, Gönen I, Kaya O, Yaylý G. The determination of susceptibility of beta-lactam antibiotics and extended spectrum beta-lactamase production in Enterobactericeae which responsible from nosocomial infections. Med. J. Sdu. 2004; 11:6-9. https://dergipark.org.tr/en/pub/omujecm/issue/20393/215902
- Turner PJ. Extended-spectrum b-lactamases. Clinical Infectious Diseases. 2005; 41(Supplement_4):S273-S275.
Crossref
© The Author(s) 2021. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.