ISSN: 0973-7510
E-ISSN: 2581-690X
There are wide range of biopolymers synthesized by diverse group of bacteria, among them intracellular polyhydroxyalkanoates (PHA) is on the priority list based on its higher level of uses and extensive studies on the processes involved in its biosynthesis. This study focused on screening of indigenous bacterial strains for PHA production. Twenty-six different indigenous bacterial strains have been inventoried and exploited for biopolymer production. The screened bacteria stained bluish-black to purple colonies upon staining with Sudan Black B indicating their potency for PHA production. The inclusion bodies produced strong orange fluorescence with staining by Nile Blue A which were further confirmed by microscopic examinations. The size distribution of PHA granules ranged from 0.5 to 1.0 ìm with the mean value of 0.5 ± 0.06 ìm. Out of 26 strains, Bacillus sp. Strain-6 and Pseudomonas sp. Strain-16 has been recognized as a potential candidate for biopolymer production and further identified through 16S rRNA gene analyses. The PHA yield of the two potent bacterial isolates being 0.84 and 1.12 g/L, and recorded 55.4 and 71.1% yield of PHA in cell dry weight (CDW), respectively. FT-IR Spectroscopic analysis of biopolymer produced by the two strains revealed two main absorptions peaks at C–H and carbonyl stretching bands characteristic to PHA. The H1 and C13 NMR spectra confirmed the presence of -CH- group in PHA extracted from the two strains.
Indigenous bacteria, Biopolymer, Sudan B black B, FT-IR spectroscopy
The biological resources have been exploited since ancient human history for potential source of substances utilizable for improvement of mankind, health and environment. During the last few decades research and development for production of biodegradable polymers has gained momentum owing to their positive attributes including biodegradability. A wide range of bacteria can produce biodegradable polymers suitable for numerous applications. Polyhydroxyalkanoate (PHA) is a group of naturally occurring biodegradable polymers of which PHB is the most common, produced by number of micro-organisms and with properties similar to conventional plastics1,2,3. Owing to its resemblance with petroleum derived plastics, PHAs have been one of the most extensively studied polymers for its potentiality in wide scale applications4. PHA has captivated a lot of profit-oriented and study attention due to its biodegradability, biocompatibility, chemical heterogeneity and its production from renewable carbon resources5-7. They are polyesters, made by a variety of microbes, grown under varying nutrient and environmental circumstances8. These polymers, which are generally lipid in character, are collected as storage substances in the sort of motile, uncrystallised, liquid particles, permitting microbial endurance under difficult circumstances9. The number and proportion of particles, the monomer arrangement, and macromolecular shape and physic- chemical attributes differ; rely on the producer organism10. They can be found within cells as light-refracting particles or as electron luminescent structures that, in overproducing mutants, give rise to a noticeable modification of the bacterial appearance11.
A vast category of bacteria namely; gram positive, gram negative and certain members of Archaea, synthesize polyhydroxyalkanoates. So far there have proposed more than 150 hydroxy alkanoic acids: Saturated, Unsaturated, Halogenated and Aromatic, which are included into the side chain of PHA and in rotation change their physical attributes, resulting them to be used in new technological practices. This differentiation is based on the variety of substrate, the polymerization nature and diverse metabolic routes involving the production of monomers supplied. The molecular mass of PHA is about 50-100 kDa, based on the nature of the polymer12 .
The contribution of the substrate monomer and the polymerization of these monomers are the two important steps implied in the biosynthesis of PHAs. The PHA synthesized by microbes relies mainly on the carbon source used. Carbon sources have been categorized as ‘relevant’ sources that give rise to monomers that are structurally homogenous to that specific carbon source and ‘irrelevant’ sources that give rise to monomers that are entirely divergent from the given carbon source. The reason for this variation can be clarified from the metabolic pathways operating in the microorganism.
Polyhydroxyalkanoates (PHAs) are polyesters that can be manufactured by some local bacterial strains or recombinant bacterial strains13. These biopolyesters are produced through metabolic alteration of different carbon sources. In PHA producing organisms, these polyesters are manufactured as intracellular carbon storage composites and vitality reserves [14]. Different biopolymers are already produced having diverse properties15. Among them, PHA biopolymers has been recognised as an alternative against conventional petroleum-based polymers16.
A variety of microorganisms are identified as PHA producers based on the carbon sources that they strive in. PHA producers can also be classified as: (i) Hydrocarbon Degraders as PHA Producers (ii) Halophiles as PHA Producers (iii) Photosynthetic Bacteria as PHA Producers (iv) Plant Growth Promoting Rhizobia (PGPR) as PHA Producers (v) Antibiotic Producers as PHA Producers. A restricted number of Gram-positive bacteria have been investigated in genera Bacillus, Caryophanon, Clostridium, Corynebacterium, Micrococcus, Microlunatus, Microcystis, Nocardia, Rhodococcus, Staphylococcus and Streptomyces17. To date, most PHA-accumulating bacteria were found to be Gram-negative which includes, Acinetobacter, Alcaligenes, Aphanocapsa, Aquaspirillum, Azomonas, Azotobacter, Beijerinckia, Caulobacter, Chromobacterium, Escherichia, Haemophilus, Moraxella, Nitrobacter, Photobacterium, Pseudomonas, Rhizobium, Rhodobacter, Vibrio, etc.14. As for archaea is concerned, PHA production to date, however, has been limited to haloarchaeal species, particularly the genera Haloferax, Halalkalicoccus, Haloarcula, Halobacterium, Halobiforma, Halococcus, Halopiger, Haloquadratum, Halorhabdus, Halorubrum, Halostagnicola, Haloterrigena, Natrialba, Natrinema, Natronobacterium, Natronococcus, Natronomonas and Natronorubrum18.
The aim of this study is to screen and characterize indigenous bacterial strains for PHA production. This study also evaluates the feasibility for maximum PHA production and underlining its potentiality through varied screening techniques.
Isolation of indigenous bacterial strain
Soil samples (10g each) were collected from different location (Petrol station Soil, House Soil, Garden Soil and Agriculture waste) of Dammam city in Eastern regions of Saudi Arabia and stored in polyethylene bag. Approximately 1 g of the samples was dispersed in 99 ml of sterile distilled water, agitated gently for two min for soil suspension and finally warmed at 60°C for 60 minutes in water bath. Subsequently, the liquid portion of the soil suspension was serially diluted and finally spreaded on Nutrient agar medium containing (g/L), Peptone 5, beef 3, NaCl 5 and agar 15. Inoculated Plates were incubated at 28-37°C for 24-48 hours. Colonies with morphological characteristics were isolated, subcultured for further purification. Finally, uncontaminated cultures were subcultured on nutrient agar slants and reserved at 4°C for further analysis19.
Screening of PHA production bacterial strain
Different types of biopolymer production media (M1,20 M2,21 and M3,22 were used for cultivation and PHAs production by different isolated strains, viz., M1 (g/L) Ammonium Sulfate 2.50, KH2PO4 1.50, Na2HPO4 3.50, MgSO4.7H2O 0.20, Glucose 20.00, Agar 20.00, Yeast extract traces and trace element solution (FeSO4.4H2O, CaCl2.2H2O, MnSO4.4H2O, ZnCl2 1 mM each) 1mL; M2 (g/L) Ammonium Sulfate 2.00, KH2PO4 6.67, (NH4)HPO4 4.00, MgSO4.7H2O 0.80, Glucose 20.00, Agar 20.00, and trace element solution (FeSO4.4H2O, CaCl2.2H2O, MnSO4.4H2O, ZnSO4.7H2O, CuSO4.5H2O, (NH4)Mo7O24, Na2B4O7.10H2O 1 mM each) 5mL; and M3 (g/L) Ammonium Sulfate 2.00, KH2PO4 0.50, MgSO4.7H2O 0.20, NaCl 0.10, Peptone 2.50, Glucose 20.00, Agar 20.00 and Yeast extract 20.00.
For screening of PHA accumulation inside the isolated bacteria the different assay methods were used viz., Sudan Black B, Viable Colony Assay and Nile Red A staining.
Sudan Black staining Method
In this method, bacterial isolates were inoculated in the specific PHA production media at 37°C for 24 hours. At the ending of incubation time, a thin film of bacteria was prepared and heat-fixed. Subsequently, the bacterial smear was stained with 0.3% Sudan black solution for 10 minutes, treated with ethanol for few seconds and washed gently with distilled water. As a contrast stain, the film was covered with red Safranin stain (0.5%) for 30 sec and gently rinsed with water to remove excess stain. Finally, air-dried bacterial film was examined under microscope using an oil immersion lenses. PHA particles found to be as blue-black inclusions within pink cells23,24.
Viable colony assay
In this experiment, all bacterial candidates were qualitatively analysed for PHA accumulation by using the viable colony technique of screening by applying Sudan Black Dye. For this purpose, nutrient agar plates supplied with 1% of glucose and ethanolic solution of 0.3% Sudan Black B dye, inoculated with bacterial isolates and incubated for 24 hours at 37°C. The colony stained with Sudan black dye and shows bluish-black to purple colonies indicated positive result for PHA production and colony does not stained with Sudan black shows negative result25,26.
Nile Red staining Method
In another trail to screen bacterial isolates for PHA accumulation, Nile Red staining was performed. During this process, bacterial smear was prepared, stained with Nile Red A stain for 20 minutes and finally cleansed with sterile water, left to dry and viewed under fluorescence microscope at wavelength 490 nm. PHA particles accumulating bacterial segregates exhibits bright yellowish orange color27.
Identification and characterization of PHA producing bacterial strains
For molecular identification of the potent PHAs producing bacteria the strains were identified by 16S rRNA gene analysis The strains were sent to Institute for Research and Medical Consultation (IRMC) and identified molecularly using :10 mM Top Taq PCR buffer, Q reagent, 10 mMdNTP’s, top Taq polymerase (Qiagen, Germany), forward primer, reverse primer (Applied Biosystems, Life Technologies Corporation, USA), distilled water and a loopfull of colony with the annealing temperature of 56 C/75 seconds in MyCyclerTM (Bio-rad, USA).All the amplified products were purified using PCR Purification Kit (Qiagen, Germany). BigDye Terminator Cycle Sequencing Kit (Applied Biosystems, Life Technologies Corporation, USA) was used to cycle sequence the purified amplicons and electophoresed in Genetic Analyzer 3500 (Applied Biosystems, Life Technologies Corporation, USA) using POP 7. EzTaxon tool was used to identify the 16S rRNA similarity of the isolated organisms with the many sequences of various bacteria28. Furthermore, Sequencing Analysis Software Version 5.4 (Applied Biosystems, Life Technologies Corporation, USA) was used to confirm the absence of background noise. MAFFT version 7 was used for the manual verification of the sequence similarity29. The potent PHAs producing bacterial strains as subjected to molecular identification using 16S rRNA gene analysis were Pseudomonas sp. strain-P and Bacillus sp. strain B as previously reported30.
Production of PHA biopolymer
A standard inoculum was prepared by inoculating a loop of the culture in 7.5 mL of nutrient broth medium containing: (g/L) Beef extract 3.00, Peptone 5.00, Sodium Chloride 5.00 in 250 mL conical flasks. The inoculated flasks were incubated at 37°C for 24 h. Approximately, 2% (v/v) were used as inoculum in all experiments and the pH maintained at 7.019,31. At the end of incubation period, cells were harvested by centrifugation at 10,000 rpm, – 4°C for 15 minutes and washed aseptically with sterile distilled water. The cell pellet was then dispersed in equal quantity of sodium hypochlorite (5.5% active chlorine) and incubated at 45°C for two hours. This extract was centrifuged at 8,000 rpm for 20 minutes and the pellet of PHA was rinsed with water and two times with ethanol: acetone combination (2:1). Eventually the pellet was dissolved in chloroform, shortly centrifuges at 8,000 rpm to get rid of non-dissolved debris. Finally, purified PHA was left to dry and verification of PHA yield was executed regularly by dry weight calculation (g/L)32.
Cell Dry weight (CDW) Determination
For estimation of the cell dry weight the whole cell mixture was again centrifuged at10,000 rpm for 10-15 minutes and the supernatant was discarded. The cell pellet was washed with water to get rid of residual of the media. Finally, the cell pellet was left to dry overnight at 60°C till constant weight (g/L). For determination of the (%) PHA biopolymer yield or accumulation, the following equation was applied according to32.
Characterization of the PHA biopolymer
The purified PHA was identified and characterized by employing Fourier Transform Infrared (FT-IR) spectroscopy and Nuclear Magnetic Resonance (NMR) spectroscopy. The existence and investigation of PHA polymer in dry cell material was confirmed by Fourier Transform Infrared Spectroscopy (FT-IR). Dried PHA polymer from bacterial candidate was fixed with KBr powder to form discs. Spectra were performed between500 and 4000 cm-1 using SHIMADZU -IRAFFINITY-2 – FT-IR spectrophotometer from the Shimadzu Corporation, Japan33.
Extracted PHA biopolymer from the PHA producing bacteria was distinguished by spectroscopic investigation. H1-NMR spectrum was performed on a BRUKER 850.1500200 MHz spectrometer at 30°C in CDCl3 as solvent. Although, C13-NMR spectral investigations were carried out at 125.65 MHz with the subsequent acquisition parameters: 298 k data point, 0.967 s acquisition time, recycle delay 1 s and contact time 4.50 ms34.
Screening and isolation of PHA production bacterial strain
PHA is widely distributed among prokaryotes as a carbon storage polymer35. These bacteria which can produce and accumulating such natural polymers have been reported from various environments36-38. In the present investigation a total of 26 bacterial strains were isolated locally from Eastern Project, Dammam, Saudi Arabia for investigating production of polyhydroxyalkanoates (PHAs) (Table 1). Bacterial isolates from various samples such as, Petrol station soil, House soil, Garden soil and Agricultural waste were collected. Twenty-six isolates acquired from the initial tests were followed by the viable colony staining method and were exposed to express the PHA production. Bacterial strains were maintained and screened for PHA production on different production media namely modified E2 medium, and on basal production medium specific for Bacillus, Pseudomonads and other gram-negative bacteria.
Table (1):
Bacterial strains screened for PHA production.
| Sample description | Bacterial Strain | Staining with Sudan Black B* |
|---|---|---|
| Petrol station soil | Bacillus sp. (Strain No. 1) | ++ |
| Arthrobacter sp. (Strain No. 2) | ++ | |
| Pseudomonas stutzeri (Strain No. 3) | ++ | |
| Bacillus licheniformis (Strain No. 4) | +++ | |
| Bacillus sp. (Strain No. 5) | + | |
| Bacillus sp. (Strain No. 6) | +++ | |
| Bacillus sp. (Strain No. 7) | +++ | |
| Bacillus altitudinis (Strain No. 8) | +++ | |
| House soil | Serratia liquefaciens (Strain No. 9) | ++ |
| Bacillus pumilus (Strain No. 10) | +++ | |
| Staphylococcus lentus (Strain No. 11) | ++ | |
| Bacillus cereus (Strain No. 12) | ++ | |
| Bacillus subtilis (Strain No. 13) | ++ | |
| Oligella ureolytica (Strain No. 14) | + | |
| Garden soil | Bacillus sp. (Strain No. 15) | ++ |
| Pseudomonas sp. (Strain No. 16) | +++ | |
| Staphylococcus (Strain No. 17) | ++ | |
| Pseudomonas sp. (Strain No. 18) | ++ | |
| Corynebacterium sp. (Strain No. 19) | + | |
| E. coli (Strain No. 20) | — | |
| E. coli (Strain No. 21) | — | |
| E. coli (Strain No. 22) | — | |
| Agriculture waste | Bacillus sp. (Strain No. 23) | +++ |
| Bacillus sp. (Strain No. 24) | ++ | |
| Bacillus sp. (Strain No. 25) | +++ | |
| Bacillus sp. (Strain No. 26) | ++ |
* Intensity of colour: No colour, -, light bluish-black, +, moderate, ++, good, +++
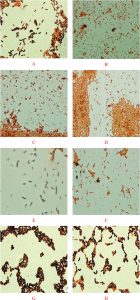
Fig. 1. Screening of PHA-producing bacteria by viable colony method and Sudan Black B staining (Magnification 100X) for Bacillus sp. Strain-6 (A,), Pseudomonas sp. Strain-16 (B), Bacillus sp. Strain-7 (C), Bacillus altitunidis Strain-8 (D), Bacillus pumilus Stain- 10 (E), Bacillus licheniformis Strain-4 (F), Bacillus sp. Strain-23 (G) and Bacillus sp. Strain-25 (H).
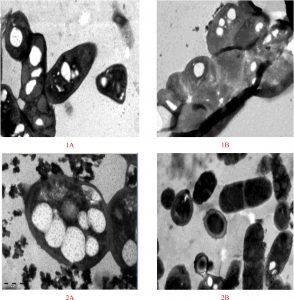
Bacterial strains stained bluish-black to purple colonies upon staining with Sudan Black B dye indicated a potent PHA producing capability, while negative PHA producers remains white, or light-blue39. PHA inclusion bodies produced strong orange fluorescence with staining by Nile Blue A.40. Results indicated that the bacterial strains-4, 6, 7, 8, 10, 16, 23, and 25 produced intensified purple colonies after the staining process (Figure 1). The results correspond to the previous findings that PHA inclusions appeared as blue-black droplets41. Indeed, these were results supported by the microscopic examination of bacterial smears from the tested organisms after staining with the same dye. Microscopic examination executed on cells comprehensibly demonstrated the presence of PHA particles within the cells. Results shown in Figure 1 indicated that out of the 26 bacterial strains Bacillus sp. Strain-6 and Pseudomonas sp. Strain-16 could be the potential candidates for PHA production. These two bacterial strains when further grown in PHA production medium showed spherical granule formation in their vegetative cells. Most of these granules were found to occupy 50% of the cells volume surrounded by compact membranes (Figure 2). The ability of Pseudomonas sp for the production of biodegradable polymers was previously reported by many researchers in their studies42-44. Bacillus sp are also noticed to be best and possible PHA accumulators in several prior investigations done by different investigators45,46. Transmission electron microscopic studies designated a better production of PHA particles within the bacteria cells in the seventy two hours trial. The results ensure the assembly of PHA by the strains. Both of the bacterial strains contained one or more PHA granules. The size distribution of PHA granules ranged from 0.5 to 1.0 µm with the mean value of 0.5 ± 0.06 µm. The bacteria cells accommodated white large inclusion particles within the cytoplasmatic fluid and also the cells were stretched and inflated based on the number of particles within the cells. Increase in the PHA accumulation may be attributed to the large cell sizes13. Comparable pictures for visualisation of PHA particles inside the bacterial cells are shown by47,48. There’s coating of phospholipids and proteins monolayer over these particles and also the linked proteins are concerned within the creation of granule and in the production and degeneration of PHA.
Fig. 2. Electron microscopic (EM) examinations of Bacillus sp. Strain-6 (1A, 1B) and Pseudomonas sp. Strain-16 (2A,2B) after growth on PAH production medium (A) as well as nutrient agar medium (B) respectively.
Quantitative determination of PHA production
In this experiment the amount of PHA produced by bacterial strains that gave bluish-black colonies during staining with Sudan Black B dye was quantitatively estimated (Table 2). For this reason, the bacterial strains cultivated on production media, polymer extracted and PHA yield was determined. Results recorded in (table 1) showed that there is great discrepancy in the amount of PHA produced by different bacterial strains. The amount of PHA produced from different strains ranged from 1.12 to 0.09 g/L which is slightly lesser than one of the previously reported research49. Results also showed that the optimal PHA production was recorded by Bacillus sp strain-6 and Pseudomonas sp. strain-16 and recorded 55.4 and 71.1% yield of PHA in cell dry weight (CDW), respectively.
Table (2):
PHA yield (%) from most potent bacterial candidates.
PHA Yield (% CDW) |
PHA (g/L) |
CDW (g/L) |
Microorganism |
|---|---|---|---|
71.1 |
0.84 |
1.19 |
Bacillus sp. Strain-6 |
55.4 |
1.12 |
2.03 |
Pseudomonas sp. Strain-16 |
17.24 |
0.18 |
0.91 |
Bacillus sp. Strain-7 |
31.82 |
0.44 |
1.38 |
Bacillus altitudinis Strain-8 |
11.54 |
0.09 |
0.82 |
Bacillus pumilus Strain-10 |
8.74 |
0.28 |
3.22 |
Bacillus licheniformis Strain-4 |
44.12 |
0.47 |
1.06 |
Bacillus sp. Strain-23 |
47.62 |
0.31 |
0.66 |
Bacillus sp. Strain-25 |
Quantitative determination of PHA production
In this experiment the amount of PHA produced by bacterial strains that gave bluish-black colonies during staining with Sudan Black B dye was quantitatively estimated (Table 2). For this reason, the bacterial strains cultivated on production media, polymer extracted and PHA yield was determined. Results recorded in (table 1) showed that there is great discrepancy in the amount of PHA produced by different bacterial strains. The amount of PHA produced from different strains ranged from 1.12 to 0.09 g/L which is slightly lesser than one of the previously reported research49. Results also showed that the optimal PHA production was recorded by Bacillus sp strain-6 and Pseudomonas sp. strain-16 and recorded 55.4 and 71.1% yield of PHA in cell dry weight (CDW), respectively.
Molecular identification and characterization of PHA producing bacterial strains
For molecular identification genomic DNA of the two potent bacterial candidates, Pseudomonas sp. and Bacillus sp. were isolated because they are the highest PHA producers among all screened isolates. Genomic DNA of these isolates was amplified using the universal primers for 16S rDNA. As per 16S RNA gene analysis by Eztaxon (Ez BioCloud), the PHA-producing bacterial candidate showed 100% similarity to P. aeruginosa type strain JCM 5962(T). Interestingly, the bacterial candidate showed 99% identity to group of P. aeruginosa strains isolated during analysis of Medium-Chain-Length polyhydroxyalkanoate producing bacteria in activated sludge samples enriched by aerobic periodic feeding (Genbank accession numbers: KY885163.1 to KY885167.1; KY885169.1 to KY885172.1; KY885174.1 to KY885176.1) Consequently, 16S RNA gene sequence was submitted to the genebank and given accession number MK027060. On the other hand, 16S RNA gene of the second bacterial candidate was amplified, sequenced and given the accession number KU199807, as reported by Berekaa et al. (2016). Fortunately, sequence analysis showed higher similarity to Bacillus aryabhattai type strain B8W22(T) and many other B. megaterium strains and was given the named B. megaterium strain DPS6. Interestingly many B. megaterium strains are known to be potent PHA producers30.
Characterization of the PHA biopolymer
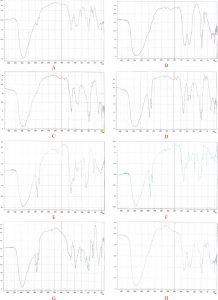
For chemical characterization of the polymer a series of spectroscopic analysis were carried out. For this purpose, a known amount of the extracted polymer was analyzed by FT-IR as well as NMR to detect the most dominant functional groups. The presence and characterization of PHA polymer in dry cell matter was confirmed by Fourier transform infrared spectroscopy (FT-IR)50. Dried PHA polymer from bacterial candidate was used to prepare KBr discs. Each chemical complex in the sample makes its own apparent alliteration to the absorbance spectrum, hence the IR spectrum of a sample represents its total chemical composition. The discreteness of an independent spectrum, that is decided by the chemical structure of every constituent and the degree to that every part contributes to the spectrum, is directly associated with the concentrations of the parts of the sample. Spectra were recorded between 400 and 4000 cm-1 using Nicolet 6700 FITR spectrometer from the Nicolet Instrument Corporation, USA. FT-IR Spectroscopic analysis of biopolymer produced by Pseudomonas sp strain-16 revealed two main absorptions peaks at C–H and carbonyl stretching bands characteristic to PHA (Figure 3). First, clear hydroxyl stretching at 3456.43 characteristic for –OH group. Absorption bands occurring at 2986.44 and 2858.50 cm-1 indicated the presence of aliphatic -CH3 and -CH2 groups. The absorption bands at 1637.56 and 1261.44 cm-1 in extracted PHA sample corresponding to the C=O and C–O stretching groups and were identical to PHA from some microbes.
Fig. 3. FTIR spectroscopic analysis of PHA biopolymer produced by Pseudomonas sp. strain-16 (A), Bacillus sp. strain-6 (B), Bacillus altitdinis strain-8 (C), Bacillus sp. strain-7 (D), Bacillus licheniformis strain-4 (E), Bacillus pumilus strain-10(F), Bacillus sp. strain-23 and Bacillus sp. strain-25
On the other hand, FT-IR Spectroscopic analysis of biopolymer produced by Bacillus sp strain P (6T) revealed two main absorptions peaks at C–H and carbonyl stretching bands characteristic to PHA (Figure 4). First, clear hydroxyl stretching at 3456.43 characteristic for –OH group. Absorption bands occurring from 2912.512 to 2989.664 and 2850.79 cm-1 indicated the presence of aliphatic -CH3 and -CH2 groups. The absorption bands from 1701.215 to 1730.147 and 1446.612 cm-1 in extracted PHA sample corresponding to the C=O and C–OH stretching groups and were identical to PHA from some microbes50-52. The results are also in accordance with the investigations on PHA recovered from Bacillus cereus and Bacillus mycoides and Bacillus thuringiensis53.
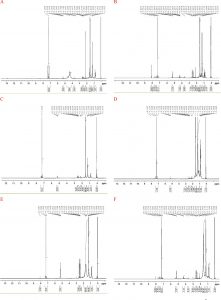
Fig. 4. H1NMR Spectroscopic analysis of PHA biopolymer produced by Pseudomonas sp. Strain-16 (A), Bacillus sp. Strain-6 (B), Bacillus altitudinis strain-8 (C), Bacillus sp. Strain-7 (D), Bacillus pumilus strain-10 (E), Bacillus sp. Strain-23 (F)
The H1 and C13 NMR were used to characterize the polymer, PHA. Signal at 5.22-5.28 in 1H spectra and corresponding signal at 68.16 in 13C spectra has confirmed the presence of -CH- group in PHA extracted from Pseudomonas sp and Bacillus sp. Signal at 0.88 in 1H spectra and corresponding signal at 169 in 13C spectra confirmed the carbonyl group in PHA. Furthermore, analysis of extracted polymer by 1HNMR revealed three groups of distinctive signals of the PHA polymer. A doublet at 1.22 and 1.25 ppm represent methyl group (-CH3) coupled to one proton and 2.28 ppm resulted from methylene group (-CH2) adjacent to an asymmetric carbon atom (Figure 4). The third signal was at 5.2 ppm attributed to a methyne group (-CH). Furthermore, C13NMR analysis was used to determine the structure of the isolated polymer from B.megaterium SW1-2 grown on the modified E2 medium. Four narrow lines appeared which were identical to the C13NMR spectra of PHA 54. The four peaks assigned for methyl (CH3, 21.2 ppm), methylene (CH2, 42.7 ppm), methine (CH, 68.5 ppm) and carbonyl (C=O, 169.7 ppm) carbon resonance of PHA 54. Analysis collectively confirmed the molecular composition of the polymer to be PHA.
Research on natural products with special emphasis on bacteria during the last decades has revealed the importance of these organisms as producers of substances useful for mankind. Recent trends in extensive burden of contaminations form conventional polymers have led to the inventorization of alternative polymers with good biodegradability. Biopolymers are the most desirable for this issue. As PHA is an alternative for plastics, it would be more useful if it is synthesized in higher concentrations. The introduction of advanced organisms to the prevailing categories of PHA-accumulating microbes shall furnish novel ways for the manufacture of economically efficient biologically degradable plastics. More recently, metabolic engineering exploiting bacterial biosynthesis pathways led to the production of new polymers with unique properties. Though PHA has been established as novel polymer in terms of their biodegradability and biocompatibility, their yield is the major constrain for their wide applicability. The present study was outlined to isolate maximum PHA producing bacterial strain among the screened isolates to obtain highest PHA yield. This study has revealed that Pseudomonas sp. and Bacillus sp. as potent isolates for the PHA production as exemplified by higher accumulation of intracellular PHA granules in their cells. The fast growth and simplicity in cultivation can result in better PHA yields in short time period. Production of PHA from indigenous bacterial strains will not only make certain reduction in production costs but will also results in reducing the environmental problems caused by waste accumulation. The characterization of PHA by different testing methods revealed the accumulation of pure PHA by the determined strains of bacteria that can be further investigated additionally by more mixing methods to obtain wider usage with improved PHA properties.
Acknowledgements
Researchers thank the research units at the Faculty of Science-Imam Abdulrahman Bin Faisal University to facilitate the performance of the practical part
Conflict of Interest
The author(s) declare that there is no conflict of interest.
- Rodriguez-Perez S., Serrano A., Alba A.P., Bernabe. Challenges of scaling-up PHA production from waste streams. Journal of Environmental Management. 2018; 205: 215-230.
- Kushwah B.S., Kushwah A.V.S., Singh V. Towards understanding polyhydroxyalkanoates and their use. J Polym Res., 2016; 23:153.
- Keshavarz T., Roy I. Polyhydroxyalkanoates: bioplastics with a green agendaþ. Curr Opin Microbiol., 2010; 13:321-326.
- Burniol-Figols A., Varrone C., Le S. Combined polyhydroxyalkanoates (PHA) and 1, 3-propanediol production from crude glycerol: Selective conversion of volatile fatty acids into PHA by mixed microbial. Water Research, 2018; 136:180-191.
- Huang L., Chen Z., Wen Q., Zhao L., Lee D., LYang. et al. Insights into Feast-Famine polyhydroxyalkanoate (PHA)-producer selection: Microbial community succession, relationships with system function and underlying. Water Research, 2018; 131:167-176.
- Anderson A, Dawes EA.Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiolþ Rev., 1990; 54:450-472.
- Shah J.P. Integrating Dry Needling with New Concepts of Myofascial Pain, Muscle Physiology, and Sensitization. Integr. Pain Med. 2008; 107–121.
- Raza Z., Abida S., Banat I.M. Polyhydroxyalkanoates: Characteristics, production, recent developments and applicationsþ. International Biodeteioration & Biodegradation., 2018; 126:45-56.
- Marang L., Loosdrecht M., Kleerebezem R. Enrichment of PHA-producing bacteria under continuous substrate supplyþ. New Biotechnology., 2018; 41:55-61.
- Zhang J., Shishatskaya E.I., Volova T.G., Ferreira Da, Silva L., Chen G.Q. Polyhydroxyalkanoates (PHA) for therapeutic applications. Materials Science and Engineering., 2018; 86:144-150.
- Naranje N.K., Wadher B.J., Purohit HJ, Muddeshwar MG. Bacillus megaterium as potential producer for Polyhydroxybutyrates. IOSR J Environ Sci., 2015; 1:7-10.
- Ushimaru K., Motoda Y., Numata K., Tsuge T. Phasin proteins activate Aeromonas caviae polyhydroxyalkanoate (PHA) synthase but not Ralstonia eutropha PHA synthase. Appl.Environ. Microbiol., 2014; 80:2867-2873.
- Chen G., Jiang X.R. Engineering microorganisms for improving polyhydroxyalkanoate biosynthesis. Current Opinion in Biotechnol., 2018; 53:20-25.
- Lu J., Tappel R.C., Nomura C.T. Biosynthesis of Poly(hydroxyalkanoates). Polym Rev., 2009; 49:226–48.
- Rehm BHA. Bacterial polymers: biosynthesis, modifications and applications. Nat Rev Microbiol., 2010; 8:578–592.
- Montiel-Jarillo G., Carrera J., Suarez-Ojeda M.E. Enrichment of a mixed microbial culture for polyhydroxyalkanoates production: Effect of pH and N and P concentrations. Science of Total Environment.2017.
- Tan G., Chen C., Li L, Ge L., Wang L., Razaad IM, et al. Start a research on biopolymer polyhydroxyalkanoate (PHA): a reviewþ.Polymers., 2014; 6:706-754.
- Han J., Hou J., Liu H., Cai S., Feng B., Zhou J,et.al. Wide distribution among halophilic archaea of a novel polyhydroxyalkanoate synthase subtype with homology to bacterial type III synthases. Appl. Environ Microbiol., 2010; 76:7811-7819.
- Singh G., Kumari A., Mittal A., Anita Y., Neeraj K.A. Poly â-hydroxybutyrate production by Bacillus subtilis NG220 using sugar industry waste water. Bio Med Research International.2013;
- Berekaa M., AlThawadi A.M. Biosynthesis of polyhydroxybutyrate (PHB) biopolymer by Bacillus megaterium SW1-2: Application of Box-Behnken design for optimization of process parameters. Afr.J.Microbiol., 2012; 6:838-845.
- Tian W., Hong K., Chen G., Wu Q., Zhang R.Q., Huang W. Production of polyesters consisting of medium chain length 3-hydroxyalkanoic acids by Pseudomonas mendocina 0806 from various carbon sources..Antonie van Leeuwenhoek. 2000; 77:31-6.
- Hamieh A., Olama Z., Hanafi H. Microbial production of polyhydroxybutyrate, a biodegradable plastic using agro-industrial waste products. Glob.Advan. J. Microbiol., 2013; 2:54-46.
- Murray R.G.E., Doetsch R.N., Robinow C.F. Determinative and cytological light microscopy, In Methods for General and Molecular BacteriologyAmerican Society for Microbiol., 1994; 21-41.
- Sharma M., Harish K.D. Isolation and optimization of culture conditions for PHB production by Bacillus megaterium. Int. J. Pharma.Bio.Sci., 2015; 6:724-734.
- Panigrahi S., Ujwala B. Screening, Isolation and quantification of PHB-producing soil bacteria. Intern.J.Engineering.Sci.Inven., 2013: 2:1-6.
- Aswini P., Kavitha P., Revathy AR, Babujanarthanam R. Poly b hydroxy butyrate (PHB) biosynthesis in Bacillus. Int J Pharm Sci Rev Res., 2014; 28:8–11.
- Bhuwal A., Singh G., Aggarwal N., Varsha G., Anita Y. Isolation and screening of polyhydroxyalkanoates producing bacteria from pulp, paper, and cardboard industry wastes. Inter.J.Biomaterials., 2013.
- Chun H., Patrick V., Deborah J. Making prudent vs. impulsive choices: the role of anticipated shame and guilt on consumer self-controlþ. Advances in Consumer Research., 2007; 34:719.
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usabilityþ. Mol. Bio. Evol., 2013; 30:772-780.
- Berekaa M., AlIssa A.M. Enhanced production of polyhydroxybutyrate (phb) from agro-industrial wastes; fed-batch cultivation and statistical media optimizationþ. J. Microbiol. Biotechnol. Food Sci., 2016; 5:606-611.
- Getachew A., Woldesenbet F. Production of biodegradable plastic by polyhydroxybutyrate (PHB) accumulating bacteria using low cost agricultural waste material. BMC Res Notes., 2016; 9:509.
- Indira M., Abraham P.K., Venkateswarulu T.C., John B.D., Nath S.B., Vidya P. Isolation, Screening and Extraction of Polyhydroxybutyrate (PHB) producing bacteria from Sewage sampleþ. Int. J. PharmTech Res., 2014; 6:850-857.
- Kansiz M., Jacobe H.B., McNaughton D. Quantitative determination of the biodegradable polymer poly (â-hydroxybutyrate) in a recombinant Escherichia coli strain by use of mid-infrared spectroscopy and Multivariative statistics. Appl. Environ Microbiol., 2000; 66:3415-3420.
- Bloembergen S, Holden DA, Hamer GK, Bluhm TL, Marchessault RH. Studies of composition and crystallinity of bacterial poly(b-hydroxybutyrate-co-b-hydroxyvalerate). Macromolecules 1986;19:2865–71.
- Nair S., Jha P.K., Babu C.R. Variation in poly-b-hydroxybutyrate synthesis in rhizobia reflects strain differentiation and temperature regulation. J Basic Microbiol., 1993; 33:35–9.
- Saito Y., Doi Y. Microbial synthesis and properties of poly (3-hydroxybutyrate-co-4-hydroxybutyrate) in Comamonas acidovoransþ. Inter.J.Biol.Mac., 1994; 16:99-104.
- Füchtenbusch B., Wullbrandt D., Steinbüchel A. Production of polyhydroxyalkanoic acids by Ralstonia eutropha and Pseudomonas oleovorans from an oil remaining from biotechnological rhamnose production. Appl Microbiol Biotechnol., 2000; 53:167–72.
- Numata K., Doi Y. Biosynthesis of Polyhydroxyalkanaotes by a Novel Facultatively Anaerobic Vibrio sp. under Marine Conditions. Mar Biotechnol., 2012; 14:323–331.
- Choi J., Lee S.Y. Factors affecting the economics of polyhydroxyalkanoate production by bacterial fermentation. Appl Microbiol Biotechnol., 1999; 51:13–21.
- López-Cortés A., Lanz-Landázuri A., García-Maldonado J.Q. Screening and Isolation of PHB-Producing Bacteria in a Polluted Marine Microbial Mat. Microb Ecol., 2008; 56:112–20.
- Arshad M., Jamil N., Naheed N., Husnain S.Analysis of bacterial strains from contaminated and non-contaminated sites for the production of biopolymersþ. Afr. J. Biotechnol., 2007; 6:1115-1121.
- Smet M. De, Eggink G., Witholt B,Wynberg HJ.Characterization of intracellular inclusions formed by Pseudomonas oleovorans during growth on octane. J. Bacteriol.1983; 154:870-878.
- Suzuki T., Yamane T., Shimizu S. Mass production of poly-b-hydroxybutyric acid by fully automatic fed-batch culture of metylotroph. Appl. Microbiol. Biotechnol., 1986; 23: 322-9.
- Aremu M.O., Layokun S.K., Solomon B.O. Production of Poly (3-hydroxybutyrate) from cassava starch hydrolysate by Pseudomonas aeruginosa. Am. J. Sci. Ind. Res., 2010; 1: 421-426.
- Full T.D., Jung D.O., Madigan M.T. Production of polyhydroxyalkanoates from soy molasses oligosaccharides by new, rapidly growing Bacillus species. Lett Appl Microbiol., 2006; 43:377–384.
- Vishnuvardhan R.S., Thirumala M., Mahmood S.K. Production of PHB and P (3HB-co-3HV) biopolymers by Bacillus megaterium strain OU303A isolated from municipal sewage sludge. World J. Microbiol Biotechnol., 2009; 25:391–397.
- Chaitanya K., Cinelli P., Lazzen A. Biotechnological production of polyhydroxyalkanoates by various isolates A Review. Int. J. Pharm Sci Invent., 2014; 3:01-11.
- Poblete-Castro I., Binger D., Oehlert R., Rohde M. Comparison of mcl-Poly(3-hydroxyalkanoates) synthesis by different Pseudomonas putida strains from crude glycerol: citrate accumulates at high titer under PHA-producing conditions. BMC Biotechnol., 2014; 14:962.
- Huang L, Chen Z, Wen Q,Lee DJ.Enhanced polyhydroxyalkanoate production by mixed microbial culture with extended cultivation strategy. Bioresource Technology, 2017; 241:802-811.
- Hong K., Sun S., Tian W., Chen G.Q., Huang W. A rapid method for detecting bacterial polyhydroxyalkanoates in intact cells by Fourier transform infrared spectroscopy. Appl. Microbiol. Biotechnol., 1999; 51:523–6.
- Pandian S., Deepak V., Kalishwarl K., Ramesshkumar N., Jeyaraj M., Gurnathan S., Optimization and fed-batch production of PHB utilizing dairy waste and sea water as nutrient sources by Bacillus megaterium SRKP-3. Bioresource Technology., 2010; 101:705-711.
- Valappil S., Misra S., Boccaccini R.A., Keshavarz T., Bucke C., Roy I., Large-scale production and efficient recovery of PHB with desirable material properties, from the newly characterised Bacillus cereus SPV. Biotech., 2007; 132:251-258.
- Rohini D., Phadnis S., Rawal S. Synthesis and characterization of poly-â-hydroxybutyrate from Bacillus thuringiensis R1. Indian J. Biotechnol., 2006; 5:276-283.
- Doi Y., Abe C. Biosynthesis and characterization of a new bacterial copolyester of 3-hydroxyalkanoates and 3-hydroxy-.omega.-chloroalkanoates. Macromolecules, 1990; 23:3705–7.
© The Author(s) 2018. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.