ISSN: 0973-7510
E-ISSN: 2581-690X
SARS-CoV-2 is highly contagious, which spreads even by patients having no clinical symptoms or also from people suffering with only mild symptoms. The gold standard test for its diagnosis is reverse-transcription PCR (RT-PCR) but at times of pandemic, Rapid antigen tests (RAT) are required, which has a very less turn-around time. Evaluation of the performance of COVID-19 Rapid antigen test in comparison to SARS-CoV-2 RT-PCR using nasopharyngeal swab, in relation to RNA dependent RNA polymerase (RdRp) Cycle threshold (Ct) values. This observational and cross-sectional study was done on patients coming with features of Influenza-like illness (ILI) or for any aerosol generating procedure or on high-risk patients seeking hospitalization. Both RT-PCR and RAT for COVID-19 were done on samples collected from each patient and results were compared. Altogether, 5314 samples were tested, out of which 104 (01.95 %) & 229 (04.31 %) samples were found positive by the RAT & RT PCR test, respectively. Sensitivity, specificity, PPV and NPV of RAT were found to be 44.54%, 99.96%, 98.08% and 97.56%, respectively. 98.9 % of samples with Ct value ≤ 20 were positive by RAT, whereas only 2.2% samples having Ct value ≥ 26 were found to be positive. Cases having lower Ct values were found to be more symptomatic and vice-versa. RAT are not efficient in detecting the virus in samples showing high Ct values (Ct ≥ 26) by RT-PCR test. Patients with samples showing low Ct values (Ct ≤ 20) had more severe symptoms and vice-versa.
SARS-CoV-2, COVID-19, Reverse Transcription Polymerase Chain Reaction, Rapid Antigen Test, RNA Dependent RNA Polymerase
The unfolding of a novel coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) causing COVID-19 infection in China, came as a disastrous surprise to the whole world in December 2019.1 The spectrum of COVID-19 infection displays a wide array of clinical conditions, ranging from a completely asymptomatic infection to severe infection requiring hospitalization or even death in some cases. SARS-CoV-2 is highly contagious, which spreads even by patients having no clinical symptoms (asymptomatic carriers) or also from people suffering with only mild symptoms.2,3 In day-to-day clinical practice, diagnosis of COVID-19 infections is done by reverse-transcription polymerase chain reaction (RT-PCR).4 The specimens for COVID-19 RT-PCR test include nasopharyngeal swabs, pharyngeal swabs, sputum, saliva, bronchoalveolar lavage, endotracheal aspirate and even stool.5
Along with the Open reading frame 1ab (Orf1ab) and the RNA dependent RNA polymerase (RdRP) gene, there are other genes which encode for N, E and S proteins, which form the targets for detection of SARS-CoV-2. The analytical specificity of RT-PCR test is affected by the type of gene under study. While there may be cross-reactivity of N gene with other coronaviruses, the E gene is specifically conserved in all beta coronaviruses. The RdRP gene is often considered as the confirmatory gene, which can be used to identify SARS-CoV-2 among all the other SARS-CoV viruses. The S gene, being highly divergent from other coronaviruses, is equally significant in differentiating SARS-CoV-2.6
Cycle threshold (Ct) value of RT-PCR is defined as the number of amplification cycles which is required by the target gene in exceeding a threshold level. Therefore, it seems that there is an inverse relation between the Ct values and viral load, which can be indirectly used to quantify the copies of viral RNA in the sample; however, the use of Ct values as an indicator of viral load is affected by multiple factors including the type of RT-PCR test used and the components of the sample matrix which may itself alter the amplification efficiency.7 All commercially available PCR kits define their range of Ct values, below which a test is considered as Positive and above which a test is considered as negative. At the same time, it has also been found that a Ct value does not correlate well with the severity of the disease.8,9
Rapid Antigen Tests (RAT) have also been devised which, in comparison to the RT-PCR tests, is very simple and can be performed in any clinical laboratory. These point-of-care (POC) tests provide tremendous benefits in patient management in respect to their reduced turn-around-time and their availability at the bedside and at remote healthcare centres. Health care providers and clinical microbiologists should be acquainted with the limitations and indications of each rapid test for their appropriate diagnostic use.10 Rapid Antigen detection tests can be of great help in COVID-19 and similar pandemics as they can be used for mass screening of a larger population in a short span of time. But at the same time, seeing the easy transmissibility of disease in COVID-19 from one person to another, any false negative results through the rapid tests will prove to be a disaster towards its containment and can cause potential surge in the cases. Also, there is paucity of data which correlate the positivity or negativity of COVID antigen test with that of the Ct value obtained by the RT-PCR test.
Aim of the study
This study was done with an aim to evaluate the diagnostic performance of COVID-19 antigen test in comparison to RT-PCR test, taking into account the RdRp Ct value obtained by the RT-PCR assay.
This observational and cross-sectional study was done on patients who came to different Departments of our college within 3 months duration (from 1st September 2020 to 15th November 2020). The study was conducted after obtaining the ethical clearance from the Institutional ethics committee.
As per the hospital policy developed during the COVID times in accordance with the ICMR guidelines,11 following patients underwent Rapid point of care test (Rapid Antigen Test) along with the gold standard RT-PCR test:
- All symptomatic *ILI (Influenza-like illness) patients suspected of having COVID-19 infection attending the hospital. (*ILI patient is defined as having acute respiratory infection along with cough and fever ≥ 38◦C).
- Asymptomatic patients attending the hospital for any surgical / non-surgical procedures having risk of aerosol generation:
- ENT surgery, neurosurgery, dental procedures etc.
- Non-surgical procedures like dialysis, bronchoscopy and upper GI endoscopy.
- Asymptomatic admitted patients or those seeking admission who belong to the outlined high-risk groups:
- Patients on chemotherapy
- Patients who are immunosuppressed
- Cancer patients
- Patients undergoing transplants.
- Elderly co-morbid patients (>65 yrs of age with pulmonary disease, cardiac disease, liver disease, renal disease, diabetes, neurological disease, blood disorders)
Nasopharyngeal swabs were collected from the patients in pairs. Rapid Antigen Tests were performed immediately under strict medical supervision as per manufacturer protocol, while samples for RT-PCR test were stored in VTM, labelled and transported to the microbiology laboratory in proper condition. Samples were then subjected to RT-PCR tests after RNA extraction within 2 hours of collection. If there was any delay in extraction procedure, samples were kept at 4°C.
Rapid antigen tests were done with different ICMR-approved kits available in the department from time to time. In rapid antigen test, nasopharyngeal swab was taken directly into buffer provided with the mentioned kit and rapid test was performed immediately as per manufacturer protocol and result was noted after 15 min and not later than 30 min of sample inoculation in the test strip well.
For RT-PCR test, nucleic acid extraction (RNA extraction) was done by using “MagNA Pure 96 DNA and viral NA Small Volume Kit” using an automated system by Roche as per manufacturer protocol. Subsequently, the RT-PCR assays were performed in Agilent AriaMx Real-time PCR system (Agilent Technologies) with the following cycling conditions: 50°C for 5 minutes for reverse transcription, one cycle each of 52°C for 5 minutes and 94°C for 10 minutes, followed by 45 cycles of 94°C for 10 seconds and 60°C for 30 seconds. One-step RT-PCR was performed to detect SARS-CoV-2 by using various ICMR-approved RT-PCR Kits, available in the department from time to time. The RT-PCR kits detected Novel coronavirus (nCOV-19) based on multiplex real-time PCR. The kits utilized primer and probes specific for nCoV-19: E gene, RdRp gene and internal control RNase P (IPC). The results of internal control signified the accuracy of sampling and nucleic acid extraction process, preventing any false negative results.
We also recorded symptoms from each individual and classified them into asymptomatic and symptomatic (mild, moderate & severe) as per ICMR guidelines:
- Mild case: Fever, symptoms of upper respiratory tract infection, SpO2 >97% on room air
- Moderate case: Symptoms of pneumonia with RR > 24/minute, SpO2< 94% on room air
- Severe case: Respiratory distress, SpO2< 90 % on room air
Inclusion criteria
Patients who were tested by both Rapid Antigen test and RT-PCR test and the records of whom were well maintained. Only single paired sample from each patient was included in the study.
Exclusion criteria
Those who underwent only one test out of Rapid Antigen test and RT-PCR test for SARS-CoV-2 and the records of whom were not well maintained. Repeat sample from a patient was also excluded from the study.
The demographic data like age & sex of each individual along with associated symptoms were recorded. The results of Rapid antigen tests were compared with the results of RT-PCR tests, considering RT-PCR as the gold standard. The detection ability of the Rapid Antigen test was studied in relation to the RdRp Ct values found in the positive RT-PCR tests.
Statistical analysis
The data after entering into MS-office Excel, were analysed using the SPSS software version 22. Descriptive statistics were used to describe general information of patients. Continuous data were presented in mean, standard deviation (SD), median, and range. Categorical data were presented in numbers, percentages, and 95% confidence interval (95% CI). An online statistical tool was used for calculating sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV).12 P value < .05 was considered for significance.
Altogether 5314 samples were tested, out of which 104 samples (1.95 %) were found positive by the Rapid Antigen test, whereas, 229 samples (4.31 %) were found positive by the RT PCR test (Table 1). Considering RT-PCR as the gold standard, the sensitivity and specificity were found to be 44.54% (95% CI, 37.99% – 51.23 %) and 99.96 % (95% CI, 99.86% – 100.00%), respectively. Positive Likelihood Ratio (PLR), Negative Likelihood Ratio (NLR), PPV and NPV are depicted in Table 2.
Table (1):
Results of RT-PCR Test and Rapid Antigen Test (n=5314)
| RT-PCR Test Result | ||||
|---|---|---|---|---|
| Positive | Negative | Total | ||
| Rapid Antigen Test Result | POSITIVE | 102 | 2 | 104 |
| NEGATIVE | 127 | 5083 | 5210 | |
| Total | 229 | 5085 | 5314 | |
Table (2):
Statistics for Rapid Antigen Test (RAT) in comparison with RT-PCR
Statistics |
Value |
95% CI |
|---|---|---|
Sensitivity |
44.54% |
37.99% – 51.23% |
Specificity |
99.96% |
99.86% – 100.00% |
Positive Likelihood Ratio |
1132.47 |
281.18 – 4561.14 |
Negative Likelihood Ratio |
0.55 |
0.49 – 0.62 |
Disease prevalence (*) |
4.31% |
3.78% – 4.89% |
Positive Predictive Value (*) |
98.08% |
92.68% – 99.52% |
Negative Predictive Value (*) |
97.56% |
97.27% – 97.82% |
Accuracy (*) |
97.57% |
97.12% – 97.97% |
(*) These values are dependent on disease prevalence.
Out of 5314 individuals tested, 2860 (53.82%) were male and 2454 (46.18%) were female. The mean age of the participants was 38.69±18.67, with a median of 38 and mode of 40. Out of all, 69 (2.41%) males and 35 (1.42%) females were found to be positive by Rapid Antigen test, whereas 157 (5.48%) males and 72 (2.93%) females were found to be positive by RT-PCR test. Among males, a maximum of 3.06% were positive by Rapid Antigen test in the age group of 41-60 years, whereas by RT-PCR test, a maximum of 6.73% were found to be positive in the age group of 21-40 years. Among females, a maximum of 1.90 % were positive by Rapid Antigen test in the age group of >60 years, whereas by RT-PCR test, a maximum of 3.38 % were found to be positive in the age group of 0-20 years (Table 3).
Table (3):
Age and Sex distribution of individuals included in this study (n=5314)
| Age distribution | 0-20 year | 21-40 year | 41-60 year | >60 year | ||||
|---|---|---|---|---|---|---|---|---|
| Gender | Male | Female | Male | Female | Male | Female | Male | Female |
| Total valid sample tested | 531 | 355 | 995 | 1079 | 847 | 758 | 487 | 262 |
| Positive by Rapid antigen | 08 (1.5%) | 0 (0%) | 27 (2.71%) | 20 (1.85%) | 26 (3.06%) | 10 (1.31%) | 8 (1.64%) | 5 (1.9%) |
| Positive by RT-PCR | 29 (5.46%) | 12 (3.38%) | 67 (6.73%) | 31 (2.87%) | 43 (5.07%) | 22 (2.9%) | 18 (3.69%) | 7 (2.67%) |
Among all RT-PCR positive results, 91 samples had Ct value ≤ 20, 48 samples had Ct value between 21 to 25 and about 90 samples had Ct value ≥ 26. On comparing the results of Rapid Antigen test and RdRp gene Ct value, we found that about 98.9 % of samples with Ct value ≤ 20 were positive by rapid antigen test (p < .05), whereas 25 % of samples with Ct value between 21 to 25 were positive (p < .05). Furthermore, only 2.2% samples having Ct value ≥ 26 were found to be positive by rapid antigen test (p < .05) (Table 4).
Table (4):
Distribution of Ct values in patients with RT-PCR positive and Rapid Antigen positive (n=5314)
Test Distribution |
Ct value ≤ 20 |
Ct value 21-25 |
Ct value ≥ 26 |
Total |
|---|---|---|---|---|
RT-PCR positive |
91 |
48 |
90 |
229 (4.31%) |
Rapid Antigen positive |
90 |
12 |
02 |
104 (1.95%) |
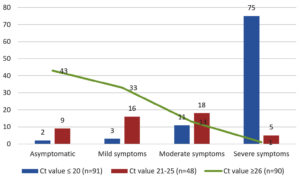
When comparing the clinical presentations among positive patients, we found only 2 (1.9%) asymptomatic cases out of 104 Rapid antigen test positive patients, whereas 54 (23.6%) asymptomatic cases were found out of 229 RT-PCR positive patients. Out of 90 cases with Ct value ≥ 26, around 43 (47.7%) cases were found to be asymptomatic. Among 175 symptomatic individuals, 81 were having severe symptoms, whereas 42 and 52 cases were having moderate and mild symptoms, respectively. About 75 (82.4%) cases with Ct value ≤ 20 were having severe symptoms (Figure).
For detection of COVID-19 cases, Real Time PCR, also known as qPCR or quantitative PCR, has been considered as the gold standard. But during the pandemic of this COVID-19, when cases started to increase at a very rapid pace, the long turn-around-time of qPCR, unavailability of specific instruments and expertise at many places became a limiting factor, as witnessed by all of us. To overcome this crisis, several Rapid Antigen tests (RAT) were licensed and used for rapid screening of cases. In the present study, we have attempted to compare the performance of RAT with respect to the qPCR method. The sensitivity of RAT was found to be around 44.54%. Sensitivity was also found to be decreased in studies done by Aleem S et al., Ristic M et al. and Lambert-Niclot S et al.,13-15 but they were more than 50%. Though low sensitivity seems to be a drawback here, less turn-around-time of RAT may be of great help at times of pandemic, where initial treatment and segregation can be started at once. There are also studies which have shown higher sensitivity like 84% and 70.6% by Igloi Z et al. and Cerutti F et al., respectively.16,17 Furthermore, the specificity of RAT in this study was found to be 99.96%, which is in accordance with the studies done by Cerutti F et al., Aleem S et al., Lee J et al. and Landaas ET et al.15-19 High specificity of a test ensures less chances of detection of a false positive case. So, if a suspected case is positive by RAT, there is high likelihood of infection due to SARS-CoV-2. In India, the point-of-care tests should be at least 50% sensitive and 95% specific for them to be accepted for use in a field setting without laboratory support.20 RAT used in our study is fulfilling the acceptance criteria for specificity, but its low sensitivity is a matter of concern.
Ct value has an inverse relationship with initial load of cDNA in case of RNA viruses like SARS-CoV-2. If the quantity of starting cDNA is more, the Ct value would be smaller and vice-versa.11 In the context of different Ct values, Binnicker21 alarmed that although real-time PCR Ct values can be used to determine the relative concentration of targeted genetic material in clinical samples, it can be impacted by many factors like sample collection, methods and kit of nucleic acid extraction, PCR amplification chemistry and the assay’s gene target(s).
Many studies have been done, correlating the Ct values of RT-PCR tests with the symptoms and viral load in an infected patient. In our study, we found a definite correlation between Ct values and disease severity, in most of the symptomatic patients. Patients with severe symptoms of disease had lower Ct values, indicating higher viral loads. Among the patients having Ct value ≤ 20, about 82.4% cases had severe symptoms, 15.4% had mild to moderate symptoms, whereas only 2.2% cases were asymptomatic. Among the patients with Ct value 21 – 25, majority of cases (70.8%) had mild to moderate symptoms, whereas about 10.4% cases had severe symptoms and 18.7% cases were asymptomatic. Among patients with Ct value ≥26, most of the cases (47.7%) were asymptomatic, whereas only 1.1% cases had severe symptoms. Many studies support the correlation of Ct values and disease severity, mortality and risk of intubation during treatment.22
In another study, it was found that mortality was around 35%, 18% and 6% among patients with a high viral load (Ct < 25), medium viral Load (Ct 25–30) and a low viral load (Ct >30), respectively. Also, the risk of intubation was around 29%, 21% and 15% in patients with a high, medium and low viral load, respectively. Zheng and Liu et al. have also reported higher viral loads and longer persistence of the virus in patients with severe disease.23,24
At the same time, there are studies in which the symptomatic and asymptomatic patients had no relation with the viral loads, as indicated by the Ct values.25 Recently, the ability of Ct values indicating the true viral load has been doubted. The Ct value of a specimen may vary in conditions where different nucleic acid extraction kits, RT-PCR kits and techniques are used. There may be variations in Ct values in different runs, even if similar kit is used in all the runs.26
When we tried to evaluate the diagnostic performance of the RAT in samples with varying Ct values as detected by their RT-PCR test, we found some astonishing results. Positive samples with Ct value ≤ 20 were also found to be positive by the RAT in about 98.9% cases (p < .05), whereas samples with Ct value between 21 to 25 were found to be positive in 25% cases (p < .05). Furthermore, samples having Ct value ≥ 26 were found to be positive only in 2.2% cases (p < .05) by rapid antigen test. Almost similar results with some differences were found in studies done by Cerutti F et al., Igloi Z et al. and Aleem S et al., which could be due to different RAT kits used in their studies.15-17 These results clearly suggests that RAT are not able to detect the disease in cases where the viral load is less (Ct value ≥ 26), but can effectively detect the cases with high viral load (Ct value < 20). At the same time, RAT may or may not detect the cases with intermediate viral load (Ct value 20 – 25). Similar concerns have been raised by other investigators too,13,14 which needs further research and improvement for a better outcome.
Limitations of the study
In this study, kits of multiple brands were used for RT-PCR and RAT. As the defined Ct value for RT-PCR tests varies from kit to kit, there may be some variations in the obtained results, which would have been more accurate if kits of a single brand were used.
The specificity of Rapid antigen test is more than the sensitivity and the negative test results must always be confirmed by the RT-PCR test. The patients whose sample shows lower Ct values (Ct < 20) are more symptomatic as compared to those whose samples show higher Ct values (Ct ≥ 26). This might be due to higher viral load in samples with lower Ct values and vice-versa. RAT methods are efficient in detecting the virus in samples with lower Ct values (Ct < 20), but are highly inefficient in detecting the virus in samples with higher Ct values (Ct ≥ 26).
ACKNOWLEDGMENTS
The authors would like to acknowledge the valuable contributions of all Lab technicians and personnel who supported and risked their lives while performing the RT-PCR tests, amidst the scary environment of COVID-19 pandemic.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
VP, KS, RiK contributed in conceptualization, literature review and data collection. VP, KS, RiK, RaK, NK & SK contributed in study design. VP, KS, RiK, RaK, NK & SK performed data analysis. VP, KS, RiK performed data interpretation. VP, KS, RiK, RaK, NK & SK wrote original draft. RaK, NK & SK reviewed the manuscript. VP, KS, RiK, RaK, NK & SK edited the manuscript. All authors read and approved the final manuscript for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This study was approved by the Institutional Ethics Committee, Indira Gandhi Institute of Medical Sciences, Sheikhpura, Patna, Bihar, India, with letter number 20/IEC/IGIMS/2021.
- Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China. N Engl J Med. 2020;382(8):727-733.
Crossref - Li R, Pei S, Chen B, et al. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV2). Science. 2020;368(6490):489-493.
Crossref - Bai Y, Yao L, Wei T, Tian F, Jin DY, Chen L WM. Presumed asymptomatic carrier transmission of COVID-19. JAMA. 2020;323(14):1406-1407.
Crossref - Corman VM, Landt O, Kaiser M, et al. Detection of 2019 novel coronavirus (2019- nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3):2000045.
Crossref - Wang W, Xu Y, Gao R, Lu R, Han K, Wu G TW. Detection of SARS-CoV-2 in different types of clinical specimens.Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020;323(18):1843-1844.
Crossref - Li C, Zhao C, Bao J, Tang B, Wang Y, Gu B. Laboratory diagnosis of coronavirus disease-2019 (COVID-19). Clin Chim Acta. 2020;510:35-46.
Crossref - Bustin SA, Mueller R. Real-time reverse transcription PCR (qRT-PCR) and its potential use in clinical diagnosis. Clin Sci. 2005;109(4):365-379.
Crossref - Indian Council of Medical Research. Advisory on Correlation of COVID-19 Disease Severity with Ct Values of the Real Time RT-PCR Test. Published August 5, 2020
- Shah S, Singhal T, Davar N, Thakkar P. No correlation between Ct values and severity of disease or mortality in patients with COVID 19 disease. Indian J Med Microbiol. 2021;39(1):116-117.
Crossref - Clerc O, Greub G. Routine use of point-of-care tests: Usefulness and application in clinical microbiology. Clin Microbiol Infect. 2010;16(8):1054-1061.
Crossref - Indian Council of Medical Research. Advisory on Newer additional strategies for COVID-19 Testing. Published June 23, 2020
- MedCalc Software Ltd. Diagnostic test evaluation calculator. https://www.medcalc.org/calc/diagnostic_test.php (Version 20.106; accessed April 12, 2022).
- Ristic M, Nikolic N, Cabarkapa V, Turkulov V, Petrovic V. Validation of the STANDARD Q COVID-19 antigen test in Vojvodina, Serbia. PLoS ONE. 2021;16(2):e0247606.
Crossref - Lambert-Niclot S, Cuffel A, le Pape S, et al. Evaluation of a rapid diagnostic assay for detection of sars-cov-2 antigen in nasopharyngeal swabs. J Clin Microbiol. 2020;58(8):e00977-20.
Crossref - Aleem S, Zahoor N, Jeelani A, Khan SS. Diagnostic Accuracy of STANDARD Q COVID-19 Antigen Detection Kit in Comparison with RT-PCR Assay using Nasopharyngeal Samples in India. J Clin Diagnostic Res. 2022;16(1):DC01-DC05.
Crossref - Cerutti F, Burdino E, Milia MG, et al. Urgent need of rapid tests for SARS CoV-2 antigen detection: Evaluation of the SD-Biosensor antigen test for SARS-CoV-2. J Clin Virol. 2020;132:104654.
Crossref - Igloi Z, Velzing J, van Beek J, et al. Clinical evaluation of roche SD biosensor rapid antigen test for SARS-CoV-2 in municipal health service testing site, the Netherlands. Emerg Infect Dis. 2021;27(5):1323-1329.
Crossref - Lee J, Kim SY, Huh HJ, et al. Clinical performance of the standard Q COVID-19 rapid antigen test and simulation of its real-world application in Korea. Ann Lab Med. 2021;41(6):588-592.
Crossref - Landaas ET, Storm ML, Tollånes MC, et al. Diagnostic performance of a SARS-CoV-2 rapid antigen test in a large, Norwegian cohort. J Clin Virol. 2021;137:104789.
Crossref - Council of Medical Research. ICMR-National Institute of Virology (ICMR_NIV), Pune Standard Operating Procedure For 2020.
- Binnicker MJ. Can the severe acute respiratory syndrome coronavirus 2 polymerase chain reaction cycle threshold value and time from symptom onset to testing predict infectivity? Clin Infect Dis. 2020;71(10):2667–8
- Magleby R, Westblade LF, Trzebucki A et al. Impact of SARS-CoV-2 Viral Load on Risk of Intubation and Mortality Among Hospitalized Patients with Coronavirus Disease 2019. Clin Infect Dis. 2021;73(11):e4197–e4205.
Crossref - Zheng S, Fan J, Yu F, et al. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: Retrospective cohort study. BMJ. 2020;369:M1443.
Crossref - Liu Y, Yan LM, Wan L, et al. Viral dynamics in mild and severe cases of COVID-19. Lancet Infect Dis. 2020;20(6):656-657.
Crossref - Zou L, Ruan F, Huang M, et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382(12):1177-1179.
Crossref - Han MS, Byun JH, Cho Y, Rim JH. RT-PCR for SARS-CoV-2: quantitative versus qualitative. Lancet Infect Dis. 2021;21(2):165.
Crossref
© The Author(s) 2023. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.