ISSN: 0973-7510
E-ISSN: 2581-690X
Alternative sources of energy are the demand of the modern world. For generating different alternate fuels, utilization of lignocellulolytic biomass is on top of the priority list, for which cellulase enzymes are required specifically. In this context, the aim of the presented paper was the partial purification and characterization of cellulolytic enzymes from Trichoderma reesei inoculated digested biogas slurry. The enzyme was produced at previously standardized conditions (Incubation period: 15 day, Spore concentration: 108 spores/ml, Slurry concentration: 25%). The cellulolytic enzymes viz. CMCase, Cellobiase and FilterPaperase produced were then partially purified by ammonium sulphate precipitation (0-30 and 30-80%) and dialysis followed by ion exchange chromatography using DEAE-cellulose column. Twelve fold purification was achieved for cellobiase. Specific activity of 20.18 U/mg was measured. Two isoforms of Cellobaise (C-I and C-II) were found with 21 and 32 fold purification, respectively. Upon characterization, the optimal pH and optimal temperature values for cellobiase came out to be 7.5 and 55°C for C-I and 25°C for C-II. Carboxymethyl cellulase was partially purified up to 10.4 fold with specific activity of 1.87 U/mg of protein and Fpase was purified to 11.3 fold with 1.47 U/mg of protein specific activity. Partially purified enzyme activities were compared with that of commercial enzymes. This is a novel work where cellulases were extracted and partially purified from digested slurry from biogas plant, which is very significant with reference to not only disposal of digested biogas slurry but also its value addition for industrial applications.
Digested Biogas Slurry, Enzymes, Ion Exchange Chromatography, Partial Purification, Trichoderma reesei.
Cellulose being the most abundant polymer on earth is considered one of the most abundant renewable sources in the world. To utilize this cellulose, it needs to be degraded to simpler sugars either with the help of acids or enxzymatically. Over the years, biological degradation of cellulose by producing cellulolytic enzymes through fungi as well as bacteria, has been extensively studied (Tomme et al, 1995). The concept of recovery of the lignocellulolytic enzymes such as cellulases (Alam et al, 2008), hemicellulase, xylanase, lignin peroxidase (Alam et al, 2009) is a new biotechnological approach for the biodegradation and biosolids accumulation of slurry beside the production of industrial enzyme which exhibit the benefit of being very low treatment and production cost and environment friendly (Alam et al, 2003).
Hydrolysis of cellulose into glucose requires a complex system of cellulolytic enzymes known as cellulases that belong to the glycosyl hydrolases family (Henrissat, 1997). There are three major groups of enzymes in cellulase system on the basis of their action: endo-b-1, 4-glucanase or non-processive cellulases (EG, EC 3.2.1.4 alternatively called Cx, CMCase and endoglucanase or 1,4-b-D-glucanohydrolase), exo-b-1, 4-glucanase or processive cellulases (avicelase, exoglucanase, cellobiohydrolase, G1, CBH, EC 3.2.1.91) and b-1, 4-glucosidase (BG, EC 3.2.1.21)Gao et al,2008; Jagtap and Rao 2005; Thongekkaew et al,2008). Endocellulases act randomly and cleave the cellulose at exposed positions and produce new reducing ends. The exocellulases remain attached to the chain and release mainly cellobiose or cellotetraose units from one end of the chain (Sakon et al, 1997). For the complete hydrolysis of cellulose into monomer glucose the synergistic action of all three cellulolytic enzymes including endo-b-1,4-glucanase (EC 3.2.1.4, EG, randomly cleaving internal linkages), cellobiohydrolase (EC 3.2.1.91, CBH, specifically hydrolyzing cellobiosyl units from non-reducing ends), and b-D-glucosidase (EC 3.2.1.21, hydrolyzing glucosyl units from cellooligosaccharides) is required (Perez et al, 2002). The cellulases are used in many industrial as well as biotechnological applications including animal feed, textile, waste water, brewing and wine-making (Beguin and Auburt, 1994).
Due to increasing energy demand, focus is now on production and purification of cellulolytic enzymes from abundant renewable lignocellulosic biomass, especially agricultural waste residues, agro-industrial waste and their by-products which can help to reduce cellulase prices (Rodriguez-Couto and Sanroman, 2005). When microorgtanisms grow on cellulosic substrates, the cellulase enzymes are induced. Many different microorganisms are able to produce cellulase enzyme including fungi, bacteria and actinomycetes. The majority of the commercial and laboratory cellulases are produced using fungi due to their high enzyme activity (Nagendran et al, 2009). Among fungi, Trichoderma species are able to produce numbers of extracellular polysaccharide hydrolyzing enzyme (Bhat and Bhat, 1997).Enough nutrients for cellulase production are present in slurry, making it an efficient substrate for enzyme production. Slurry contains considerable amount of valuable substances.
For understanding the mechanism of cellulose degradation by the enzyme, it is necessary to isolate, purify and characterize these enzymes. Variety of techniques have been used for this purpose i.e. gel filtration, ion-exchange chromatography, preparative polyacrylamide gel electrophoresis, high pressure liquid chromatography and preparative iso-electric focusing. A few workers have taken the advantage of selective binding of cellulases to their insoluble substrates i.e. crystalline cellulose, cellulose powder and natural sources of cellulose i.e. wheat straw and cotton linters. Cellobiohydrolases of T.viride has been purified by applying the same phenomena. Mart-Yanov et al (1983) has reported single step purification of cellulases by using adsorption chromatography on crystalline cellulose with 50 time purification from the original sample. Extracellular cellulose binding endoglucanase of Cellulomonas species has also been purified by affinity chromatography on phosphoric acid swollen cellulose. Lamed et al (1987) have purified and partially characterized a complete cellulase complex “cellulosome” from culture supernatant of Clostridium thermocellum by using adsorption chromatography on micro-crystalline cellulose and gel filtration on Sepharose-B.
Chen et al (2004), purified CMCase from Sinorhizobium fredii. The crude protein was first precipitated with a 40-60% ammonium sulphate saturation. Two step chromatography procedure was employed including DEAE Sepharose anion exchange and phenyl sepharose. Ahmed et al (2009) purified cellulase at 4°C by ammonium sulphate precipitation, gel filteration on Sephadex G-200 column followed by gel filteration on Sephadex G-50 column. Bara et al, (2003) purified exoglucanase (EXG) by gel filteration and ion exchange chromatography and observed 35.7 fold purification with 9.5% yields. Noronha et al (2000) has employed gel filteration and ion exchange chromatography for purification of EXG and got 69 fold purification with 0.32% yield. Pitson et al (1997) purified b-glucosidase by ammonium sulphate precipitation and gel filteration. Wei et al (1996) purified b-glucosidase by ammonium sulphate precipitation, ion exchange and gel filteration chromatography. b-Glucosidase was purified from Monascus purpureus by gel filteration chromatography with 13 fold purification (Dariot et al, 2008). b-Glucosidase was purified from Paecilomyces thermophila by gel filteration chromatography with 105 fold purification and final recovery of 21.7% (Yang et al, 2008).
Okada (1976) purified cellulase-III component from a crude cellulase preparation of Trichoderma viride by DEAE Sephadex A-50 column chromatography technique. Optimum pH was 4.5-5.0 and temperature was 50°C. Begum et al (2009) purified extracellular cellulase from Aspergillus niger ITCC-4857.01 by ion exchange chromatography using DEAE Cellulose followed by gel fileration. The purification achieved was 53 fold from the crude extract with a yield of 49%. Shanjing et al (2002) purified endoglucanase by gel filteration by Superdex-75 prep grade with an active recovery of 92.8% and the purification factor 4.2. The optimum temperature and pH for the enzyme were 55°C and 4.5-5.0, respectively.Wood et al (1972) purified C1 component of Trichoderma koningii cellulase by chromatography on DEAE Sephadex with a salt gradient.
Youssef (2011) reported specific activity by A.oryzae a maximum of 24.2,18.1 and 1.65 U/mg biomass at 40°C of endo-,exoglucanase and filer paperase activity (FPA). A temperature of 40°C was identifies as optimum for metabolite production and sugar utilization by A. niger ATCC 10577 and A.niger V Tiegham (Roukas,2000; Fawole and Odunfa, 2003).
Hence the present study is aimed to accomplish the following objective:
- To partially purify cellulolytic enzymes extracted from Trichoderma reesei MTCC 164 inoculated digested biogas slurry.
Source of substrates, chemicals and microbial culture
Digested biogas slurry was obtained from a working biogas plant in biogas field laboratory of School of Renewable Eneergy Engineering, PAU, Ludhiana. The analytical grade chemicals required for media and solutions preparation were purchased from Hi-Media, SRL, Sigma and S.D fine chemicals Pvt. Ltd. Standard culture of Trichoderma reesei MTCC 164 was procured from MTCC (Microbial Type Culture Collection), Institute of Microbial Technology, Chandigarh and was maintained on malt extract Blakeslee‘s agar (composition given in Annexure I) slants at 30±20C by monthly transfers. The culture was stored in refrigerator after sub-culturing.
Production of Cellulolytic Enzymes
Digested slurry was inoculated with 2ml spore suspension of T. reesei spores @ 10v spores/ml and was incubated at 30±2°C for enzyme production. After 10 days, crude enzyme was extracted by centrifugation and supernatant was analyzed for activities of carboxymethylcellulase, filterpaperasee and cellobiase by Mandels et al (1976) method. Total protein content was determined by the method of Lowry et al, (1951).Enzyme activities (U/ml of sample) and protein (mg/ml of sample) was determined spectrophotometrically using UV-VIS spectrophotometer 2800 model. The carboxy methyl cellulase (CMCase) activity was measured according to method of Mandels et al (1976). The cellobiase activity was measured by the methods of Toyama and Ogawa (1977). FPase activity was measured according to method of Mandels et al (1976).
Enzyme units
The cellulase enzyme activity is expressed in terms of Internatinal units and international unit of cellulase may be defined as 1 micromole of reducing sugar released per minute per mililitre of enzyme extract, measured as glucose. Appropriate dilution factors were used as and when followed during estimation of enzyme activity
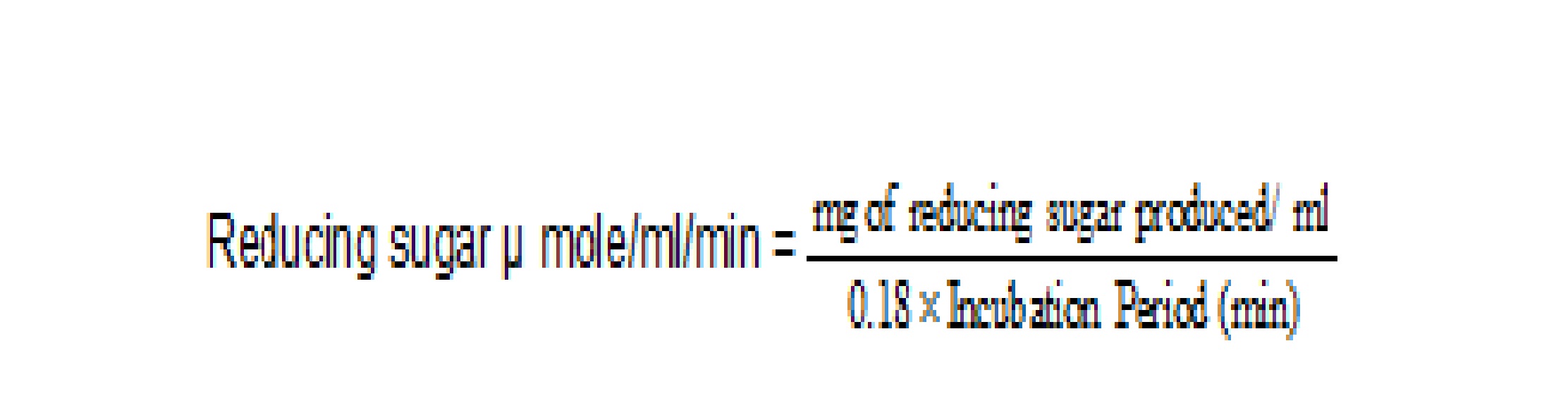 Partial Purification of Cellulolytic Enzyme from T. reesei Inoculated Digested Biogas Slurry
Partial Purification of Cellulolytic Enzyme from T. reesei Inoculated Digested Biogas Slurry
Crude enzyme extraction and enzyme essay
The crude enzyme was produced under already optimized condition (i.e. 25% concentration of slurry, 10x spores/ml spore concentration and 15 day incubation) from previous experiments and was centrifuged at 10,000 rpm for 15 min at 4°C. The supernatant was used as crude enzyme extract.
Precipitation with (NH4)2SO4 and dialysis
The crude enzyme produced was subjected to precipitation at 0-30% and 30-80% ammonium sulphate saturation. The precipitates were then dissolved in minimum volume (10-20ml) of 0.1M phoshphate buffer (pH 6.0) and dialyzed against excess of diluted (5x) buffer for 48 hrs at 4°C. Activity of cellulases was tested in the protein dialysate and was further purified by DEAE-Cellulose column chromatography.
Enzyme purification by DEAE-Cellulose column chromatography
Activation of DEAE-cellulose
Excess of 0.5N HCl (10ml) was added to about 25 g of DEAE-cellulose, mixed nicely, kept for 30 minutes and supernatant was removed slowly. This procedure was repeated five times followed by the washings with distilled water 3 times. Then sufficient amount of 0.5N NaOH was added to above treated DEAE cellulose in the same way five times and washed with distilled water so that maximum DEAE cellulose had settled down and its colour became white. Before packing into the column DEAE cellulose was given washings with 0.1M phosphate buffer (pH 6.0) and then slurry of activated DEAE-cellulose was poured in the column (shown in Fig.1) without interruption. The column was allowed to settle down uniformly and was equilibrated with phosphate buffer (pH 6.0) until the eluate showed a pH of 6.0.
 Fig. 1: Activation of DEAE cellulose column and ion exchange chromatography
Fig. 1: Activation of DEAE cellulose column and ion exchange chromatographyEnzyme purification
The crude extract was subjected to purification after dialysis at 4°C. The protein dialysate was loaded on DEAE cellulose column equilibrated with 0.1M phosphate buffer (pH 6.0). The enzyme was eluted with same buffer using stepwise gradient (0.1M each) of increasing molarity of NaCl (0.1-1.0 M) at a flow rate of 12ml per hour (as shown in Fig 1). 25ml buffer of each molarity was used for elution. Fractions of 5ml each were collected and analyzed for cellulase activity and protein content. The fractions which contained the sufficient high activity of cellobiase were separately pooled for further characterization.
Characterization of Cellobiase
Determination of optimum temperature
Activity of cellobiase was determined at different temperatures ranging 5-35°C. The value of optimum temperature was determined by plotting percent maximum activity verses temperature.
Determination of optimum pH
Activity of cellobiase was determined at different pH ranging from 5.8 to 8.0. The value of optimum pH was determined by plotting percent maximum activity verses pH.
Statistical Analysis
Standard error was calculated manually for all the experiments. All treatments were completed in triplicate.
The present study was conducted to partially purify cellulolytic enzymes from digested biogas slurry using a mesophilic, filamentous fungi Trichoderma reesei MTCC 164.
Partial Purification of Cellulolytic Enzyme from Digested Biogas Slurry Using T.Reesei MTCC 164
The crude enzyme extract was initially analyzed for all cellulolytic enzyme activities. Results from Table 1 indicate that the extract showed maximum Cbase activity i.e. 3406.48 U/ml, followed by Fpase activity (428.57 U/ml). The crude enzyme extract was subjected to precipitation with 0-80% ammonium sulphate saturation in two steps i.e. 0-30% and 30-80%. Dialysis was performed with both precipitate samples. Precipitates of 30-80% saturation showed maximum activities after dialysis i.e. 29.92 U/ml of CMCase, 284.12 U/ml of Cbase and 20.68 U/ml of Fpase. Protein content was measured to be 169.28 mg/ml and reducing sugars to be 475.41 mg/ml.
Table (1):
Partial purification of cellulolytic enzymes upto dialysis from digested biogas slurry using T. reeseiMTCC 164.
Sample |
CMCase activity (IU/Kg digested slurry) |
Cbase activity (IU/Kg digested slurry) |
Fpase activity (IU/Kg digested slurry) |
Protein content (mg/l of digested slurry) |
Reducing Sugars (mg/l of digested slurry) |
|---|---|---|---|---|---|
Units in crude extract |
321.53 ± 9.30 |
3406.48 ± 31.56 |
428.57 ± 11.4 |
2610.4 ± 14.6 |
1512.2 ± 11.8 |
After 0-30% (NH₄)₂SO₄ precipitation |
21.89 ± 0.56 |
172.13 ± 2.21 |
19.08 ± 0.12 |
301.47 ± 6.36 |
413.60 ± 9.48 |
After 30-80% (NH₄)₂SO₄ precipitation |
47.31 ± 0.76 |
289.12 ± 4.78 |
20.85 ± 0.49 |
522.14 ± 14.86 |
951.07 ± 16.51 |
0-30% ppt dialysate |
18.33 ± 0.06 |
136.27 ± 2.81 |
13.31 ± 0.02 |
84.82 ± 4.32 |
338.4 ± 5.67 |
30-80% ppt dialysate |
29.92 ± 0.12 |
284.12 ± 9.51 |
20.68 ± 0.06 |
169.28 ± 5.40 |
475.41 ± 11.31 |
#Cultural conditions: Amount of slurry used: 1000ml; Incubation period: 15 days; Incubation temperature: 30±2◦C; Spore concentration: 10⁸ spores/ml of suspension; Slurry concentration: 25%. The data represents the mean of three determinations each; ±values indicate standard error.
Partial purification of Cellobiase
The crude enzyme extract after 30-80% ammonium sulphate precipitation was dialyzed. Purification achieved with this step was 1.29 fold as the specific activity of enzyme increased from 1.3 IU/mg of protein in crude enzyme to 1.68 IU/mg proteins in ammonium sulphate precipitated enzyme. After anion exchange chromatography on DEAE cellulose column two different cellobiase isoforms named C-I and C-II were identified with purification folds of 21.16 and 31.83, respectively. These isoforms were eluted at 0.4-0.5 M and 0.8- 0.9 M (C I- Fraction 21 to 28 or 0.4a to 0.5c; C II- Fraction 41 to 46 or 0.8a to 0.9a) of NaCl concentrations and named on the basis of elution with increasing molarity of NaCl. The purification profile is shown in Table 2 and Fig2.
Table (2):
Purification profile of Cellobiase after Ion Exchange Chromatography.
Purification step |
Total Enzyme units produced (Units) |
Protein content (mg) |
Specific activity (units/mg of protein) |
Purification fold |
Yield(%) |
|---|---|---|---|---|---|
Units in crude extract |
3406.48±31.56 |
2610.4± 14.6 |
1.3 |
1 |
100 |
Crude Cellobiase (30-80% dialysate) |
284.119±9.51 |
169.28±5.40 |
1.68 |
1.29 |
100 |
DEAE Cellulose Cellobiase |
258.02±7.20 |
12.801±0.09 |
20.16 |
12 |
90.81 |
C-I Cellobiase |
44.226±0.36 |
1.244±0.02 |
35.55 |
21.16 |
15.57 |
C-II Cellobiase |
30.86±0.21 |
0.577±0.005 |
53.48 |
31.83 |
10.86 |
#Cultural conditions: Amount of slurry used: 1000 ml; Incubation period: 15 days; Incubation temperature: 30±2◦C; Spore concentration: 108 spores/ml of suspension; Slurry concentration: 25%. The data represents the mean of three determinations each; ±values indicate standard error.
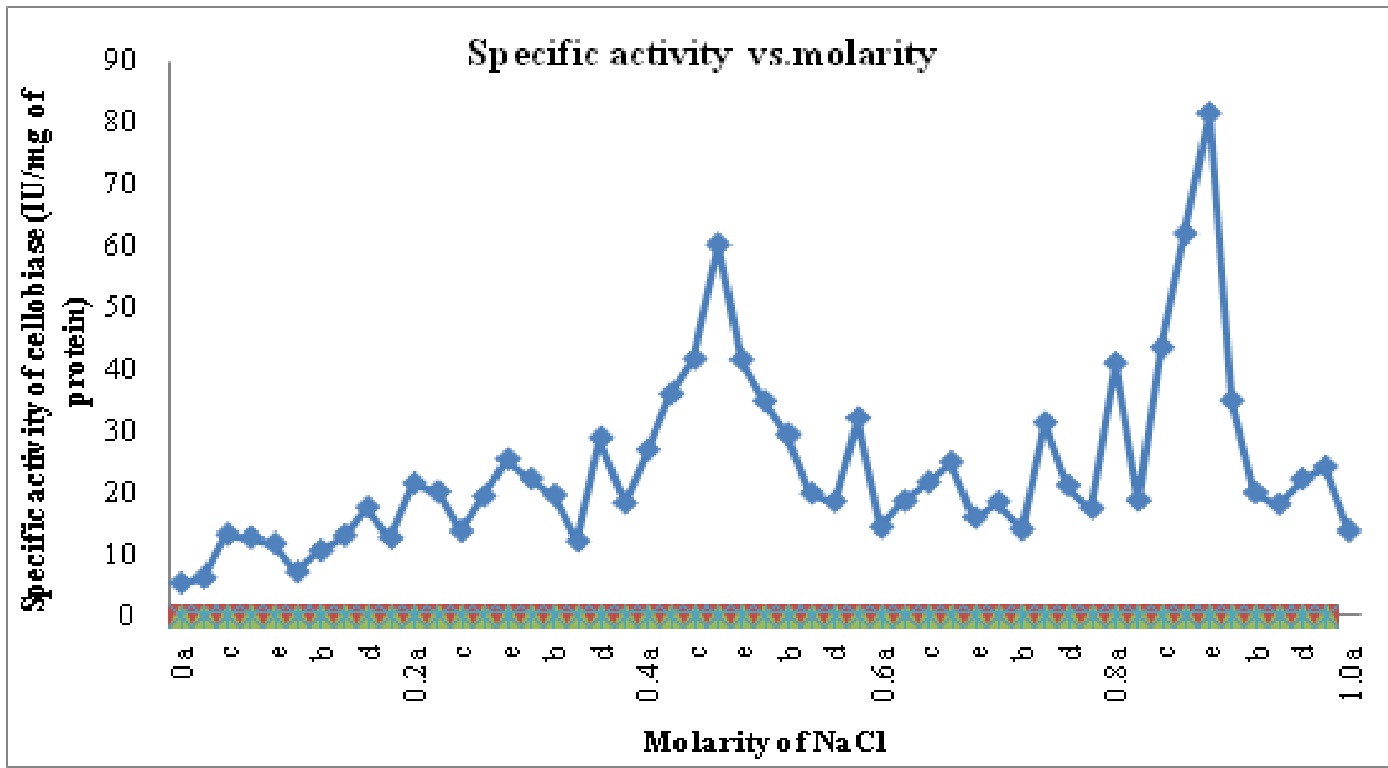 Fig. 2: Specific activity vs. molarity chart for cellobiase
Fig. 2: Specific activity vs. molarity chart for cellobiase
Shanjinget al (2002) purified cellulase by gel filtration using superdex 75 prep grade with an activity recovery of 92.8% and the purification factor 4.2. The optimum pH and temperature of purified cellulase was reported as 4.5-5.0 and 55°C, repectively. Lau and Wong (2001) purified a novel cellobiase Cba 2 from Cellulomonasbiazotea over 260 fold using ammonium sulphate precipitation, gel filtration chromatography, anion-exchange chromatography and reversed phase high performance liquid chromatography sequentially. Purified enzyme has a pH optimum of 4.8 and the optimum temperature for the activity was 70°C. Ghoraiet al (2010) purified cellobiase from the culture medium by ultra filtration and gel permeation, ion-exchange and high performance liquid chromatography. Enzyme had temperature and pH optima at 45°C and 5.4 respectively and retained full activity in a pH range of 5.0-8.0 and temperature of 30-60°C.
Effect of temperature on partially purified cellobiase
The most important factor among all the physical variables affecting the production of enzymes is usually the temperature because enzyme activities are sensitive to temperature (Krishna, 2005). Here, Cellobiase activities were measured in partially purified cellobiase at different temperatures ranging from 5 to 35°C. C-I showed maximum Cbase 3.89 units at 55°C and comparable units (3.091) at 25°C and C-II produced 3.169 units at 25°C, which is thus optimum temperature as shown in Table 3. Duttaet al (2008) studied cellulase from Penicillliumcitrinum MTCC 6489 and found maximum FPase and CMCase at 65 and 60°C respectively. Xin and Geng (2010) showed an optimum activity at 30°C for FPase and CMCase of Trichoderma reesei RUT C-30.
Table (3):
Optimum temperature of partially purified cellobiase.
Temperature (°C) |
Cbase activity in C-I (U/ml) |
Cbase activity in C-II (U/ml) |
|---|---|---|
5 |
1.739 ± 0.012 |
2.114 ± 0.017 |
15 |
2.147 ±0.009 |
2.737 ± 0.015 |
25 |
3.091 ± 0.015 |
3.169 ± 0.023 |
35 |
2.697 ± 0.021 |
2.427 ± 0.012 |
45 |
2.358 ± 0.019 |
1.479 ± 0.010 |
55 |
3.89 ± 0.024 |
2.344 ± 0.033 |
The data represents the mean of three determinations each; ±values indicate standard error.
Effect of pH on partially purified cellobiase
Cellobiase activities were measured in partially purified cellobiase at different pH ranging from 5.8 to 8. Maximum Cellobiase activity (2.197 Units/ml in Pool-I and 2.729 Units/ml in Pool-II) was observed at pH 7.5 as shown in Table 4. Maximum CMCase and cellobiase activities from Penicilliumcitrinum MTCC 6489 were reported at pH at 5.5 and 8.0 and FPase at 6.5 (Duttaet al, 2008). Xin and Geng (2010) working with horticultural waste as the substrate for cellulase and hemicellulase production from Trichoderma reesei RUT C-30 under SSF, obtained maximum cellulase at pH 5.0.
Table (4):
Cellobiase characterization: Optimum pH of partially purified cellobiase.
pH |
Cbase activity in pool-I (U/ml) |
Cbase activity in pool-II (U/ml) |
|---|---|---|
5.8 |
1.194 ± 0.001 |
1.227 ± 0.009 |
6 |
1.376 ± 0.017 |
1.535 ± 0.012 |
6.5 |
1.499 ± 0.011 |
1.553 ± 0.010 |
7 |
1.779 ± 0.009 |
1.831 ± 0.021 |
7.5 |
2.197 ± 0.018 |
2.729 ± 0.025 |
8 |
1.723 ± 0.017 |
1.302 ± 0.006 |
Comparable pH and temperature optima have been reported for cellulases of Trichoderma viride and other fungi (Berghem and Pettersson, 1973; Okada, 1975; Berg and Pettersson, 1977). Das et al (2008) reported that cellulase activity was optimum at pH 4.8. The variation of pH from the optimum level causes denaturation of the enzyme and reduces enzyme synthesis ability.
Roberto et al (2005) revealed that the highest levels of cellulase and xylanase enzymes from Thermoascusaurantiacus strain and optimum pH and temperature for cellulase and xylanase production were 5.0-5.5 and 75ºC respectively.
Partial purification of Filter Paperase
The crude enzyme extract after 30-80% ammonium sulphate precipitation was dialyzed. Purification achieved with this step was 0.8 fold as the specific activity of enzyme decreased from 0.16 in crude enzyme to 0.13 U/mg proteins in ammonium sulphate precipitated enzyme. But after anion exchange chromatography on DEAE cellulose column two different Fpase isoforms named F-I and F-II were identified with purification folds of 26.65 and 29.15, respectively. These isoforms were eluted at 0.6-0.7 M and 0.8 M (F I- Fraction 31 to 37 or 0.6a to 0.7b, F II-Fraction 39 to 44 or 0.7d-0.8d).of NaCl concentrations and named on the basis of elution with increasing molarity of NaCl. The purification profile is shown in Table 5.
Table (5):
Purification profile of FPase after Ion Exchange Chromatography.
Sample |
Enzyme units produced(Units/ml) |
Protein content(mg/ml) |
Specific activity (enzyme unit/ mg of protein) |
Purification fold |
Yield(%) |
|---|---|---|---|---|---|
Units in crude extract |
428.57 ± 11.40 |
2610.4 ± 14.60 |
0.16 |
1 |
100 |
Crude FPase (30-80% dialysate) |
20.678 ± 0.06 |
169.28 ± 5.40 |
0.13 |
0.8 |
100 |
DEAE Cellulose FPiase |
18.839 ± 0.16 |
12.801 ± 0.09 |
1.47 |
11.3 |
91.1 |
F-I FPase |
3.332 ± 0.003 |
0.962 ± 0.002 |
3.464 |
26.65 |
0.16 |
F-II FPase |
3.104 ± 0.001 |
0.819 ± 0.001 |
3.790 |
29.15 |
0.50 |
#Cultural conditions: Amount of slurry used: 1000ml; Incubation period: 15 days; Incubation temperature: 30±2◦C; Spore concentration: 108 spores/ml of suspension; Slurry concentration: 25%; The data represents the mean of three determinations each; ±values indicate standard error.
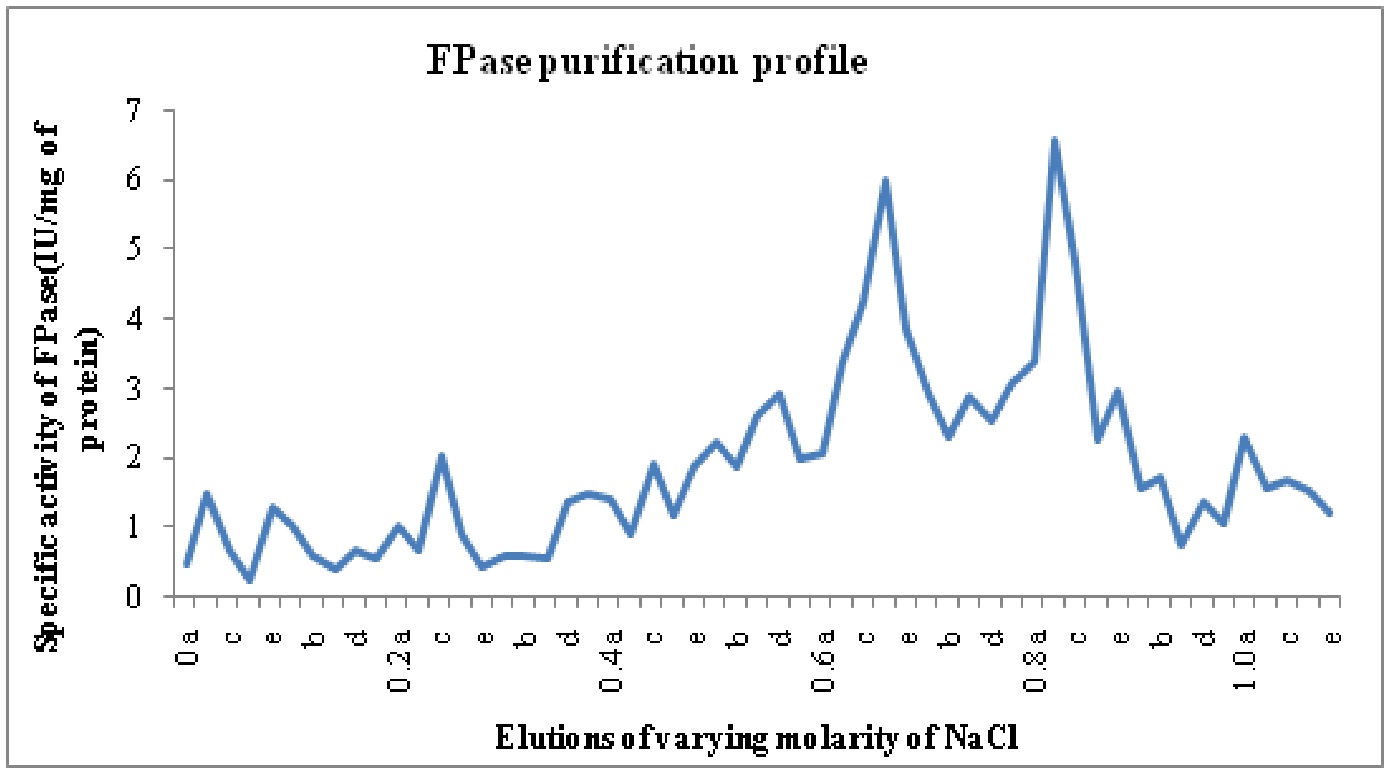 Fig. 3: Specific activity vs. molarity chart for filterpaperase
Fig. 3: Specific activity vs. molarity chart for filterpaperase
Partial purification of Carboxymethylcellulase
The crude enzyme extract after 30-80% ammonium sulphate precipitation was dialyzed. Purification achieved with this step was 1.5 fold as the specific activity of enzyme increased from 0.12 in crude enzyme to 0.18 U/mg proteins in ammonium sulphate precipitated enzyme. After anion exchange chromatography on DEAE cellulose column two CMCase isoforms namely CMC-I and II was identified with purification folds of 15.2 and 8.3. The isoforms were eluted at 0.8-0.9 M (POOL I- Fraction 43 to 47 or 0.8c to 0.9b) and 0.3 M (Pool II- Fraction 16-20) of NaCl concentration and named on the basis of elution with increasing molarity of NaCl. The purification profile is shown in Table 6.
Table (6):
Purification profile of CMCase after Ion Exchange Chromatography.
Sample |
Enzyme units produced(Units/ml) |
Protein content (mg/ml) |
Specific activity (enzyme unit/mg of protein) |
Purification fold |
Yield (%) |
|---|---|---|---|---|---|
Units in crude extract |
321.53 ± 9.30 |
2610.4 ± 14.60 |
0.12 |
1 |
100 |
Crude CMCase (30-80% dialysate) |
29.92 ± 0.12 |
169.28 ± 5.40 |
0.18 |
1.5 |
100 |
DEAE-Cellulose CMCase |
23.93 ± 0.11 |
12.801 ± 0.09 |
1.87 |
10.4 |
79.9 |
Pool-I CMCase |
2.475 ± 0.01 |
0.904 ± 0.002 |
2.74 |
15.2 |
8.27 |
Pool-II CMCase |
2.475 ± 0.01 |
1.296 ± 0.007 |
1.5 |
8.3 |
6.43 |
#Cultural conditions: Amount of slurry: 1000ml; Incubation period: 15 days; Incubation temperature: 30±2◦C; Spore concentration: 108 spores/ml of suspension; Slurry concentration: 25%; The data represents the mean of three determinations each; ±values indicate standard error.
Roy et al (2005) reported that biological activities of urea denatured forms of carboxymethylcellulase, cellobiase and β-glucosidase can be recovered using three phase partitioning using (NH4)2 SO4 and t-butanol.
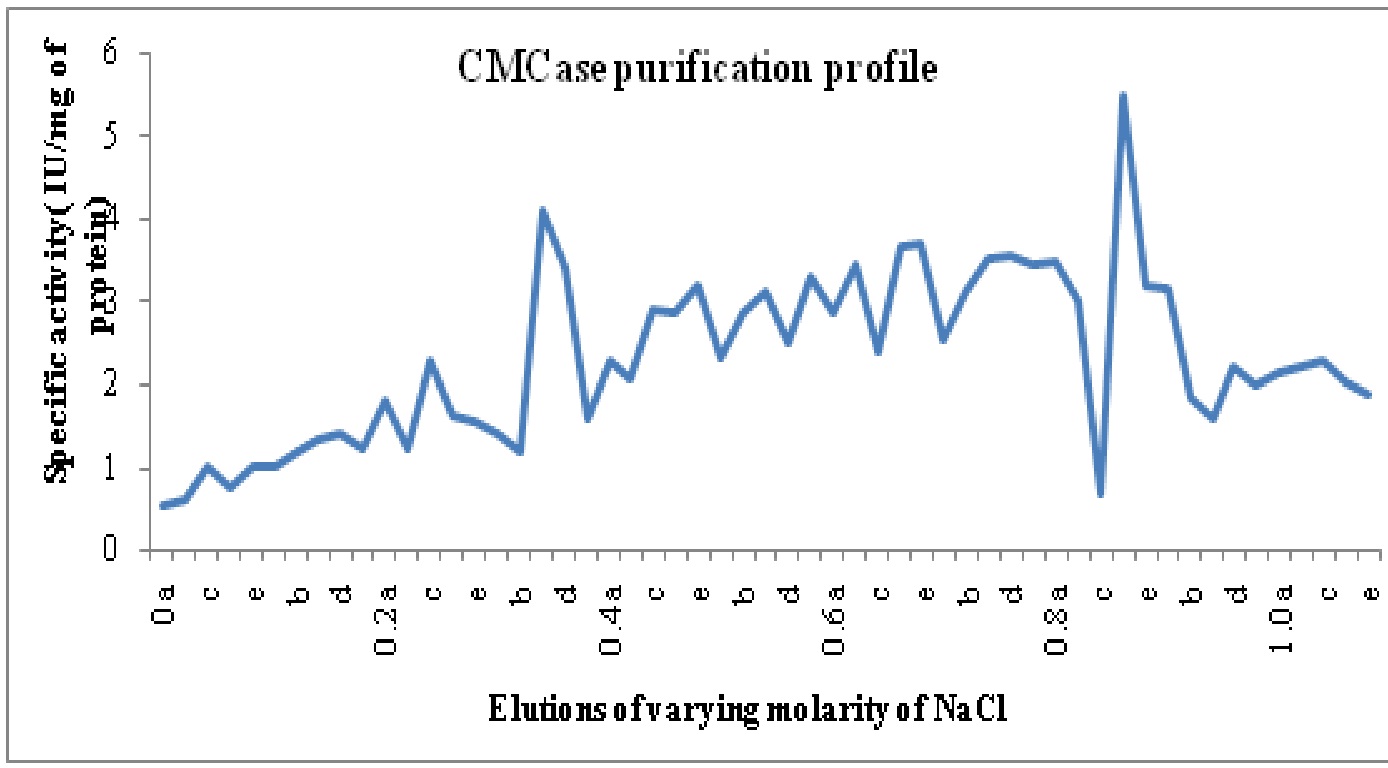
Fig. 4: Specific activity vs. molarity chart for carboxymethylcellulase
Comparative Evaluation of Partially Purified and Commercially Available Enzyme
Partially purified cellulase was compared with commercial cellulase from A. niger as shown in Table 7. The specific activities of all the three components of cellulase complex were very less as compared to commercial enzyme. This might be due to incomplete purification of enzyme in laboratory conditions from Trichoderma reesei.
Table (7):
Specific activities of commercial and partially purified enzyme.
Enzyme type |
CMCase (U/mg of protein) |
Cbase(U/mg of protein) |
Fpase(U/mg of protein) |
|---|---|---|---|
Commercial cellulase from A. niger(SISCO Lab. pvt. ltd.) |
23.15 Worth 1307 Rs/100mg |
169.86 |
16.28 |
Partialy purified Cellulase from T. reesei (Biogas Lab, PAU) |
1.86 |
1.57 |
1.47 |
The present study was aimed at partial purification of cellulolytic enzymes from digested biogas slurry using celluloytic fungi Trichoderma reesei MTCC 164. From the previous laboratory studies, it was reported that statistically the optimized conditions for maximum cellulolytic enzyme production from digested biogas slurry are: Incubation period: 15 day, Spore concentration: 108 spores/ml, Slurry concentration: 25%. Under these conditions, Cellulolytic enzymes viz. CMCase, Cellobiase and FilterPaperase were produced and partially purified by ammonium sulphate precipitation (0-30 and 30-80%) and dialysis followed by ion exchange chromatography using DEAE-cellulose column. Twelve fold purification was achieved for cellobiase. Specific activity of 20.18 U/mg was measured. Two isoforms of Cellobaise (C-I and C-II) were found with 21 and 32 fold purification, respectively. Partially purified cellobiase was characterized for optimal pH and optimal temperature values. Optimum pH came out to be 7.5 and optimum temperature came out to be 55°C for C-I and 25°C for C-II. Carboxymethyl cellulase was partially purified up to 10.4 fold with specific activity of 1.87 U/mg of protein and Fpase was purified to 11.3 fold with 1.47 U/mg of protein specific activity. Partially purified enzyme activities were compared with that of commercial enzymes. This is the first report where cellulases were extracted and partially purified from digested slurry from biogas plant, which is very significant with reference to not only disposal of digested biogas slurry but also its value addition for industrial applications. It was noticed that further purification can be achieved by Gel filtration, Sephadex-G-50, Sephadex-G-200 column chromatography techniques. Further research is required to get insight into the further purification of partially purified cellulolytic enzymes to match up with commercial enzyme production, using gel filteration, Sephadex-G-50 and G-200 columns. The supernatant left after ammonium sulphate precipitation, can be utilized as field nutrient. It is concluded that the whole process of enzyme production from cattle dung slurry is not only a value addition of so called environmental menance, but also will be a profitable venture at commercial scale.
- Ahmed, S., Bashir, A., Saleem, H., Saadia, M., Jamil, A. Production and purification of cellulose degrading enzymes from a filamentous fungus Trichoderma harzianum. Pak J Bot, 2009; 41: 1411-19.
- Alam, M. Z., Mansor, M. F., Jalal, K.C.A., Optimization of decolorization of methylene blue by lignin peroxidase enzyme produced from sewage sludge with Phanerochaete Chrysosporium. J Hazardous Mater, 2009; 162: 708-15.
- Alam, M.Z., Razi, A.F., Molla, A.H., Biosolids accumulation and biodegradation of domestic wastewater treatment plant sludge by developed liquid state bioconversion process using a batch fermentor. Water Res, 2003; 37: 3569-78.
- Alam, Z.M., Muyibi, S.A.,Wahid, R., Statistical optimization of process conditions for cellulase production by liquid state bioconversion of domestic wastewater sludge. Bioresour Technol, 2008; 99: 4709-16.
- Bara, M. T. F., Lima, L., Ulhoa, C.J., Purification and characterization of an exo-b-1,3-glucanase produced by Trichoderma asperellum. FEMS Microbiol Lett, 2003; 219: 81-85.
- Beguin, P., Aubert, J. P., “The biological degradation of cellulose,”FEMS Microbiology Reviews, 1994; 13(1): 25-58
- Begum, F., Absar, N., Alam, M.S., Purification and characterization of extracellular cellulase from Aspergillus oryzae ITCC-4857.01. J Appl Sci Res 2009; 5(10): 1645-51.
- Begum, M. F., Absar, N. Purification and characterization of intracellular cellulase from Aspergillus oryzae ITCC-4857.01 Mycobiol 2009; 37(2): 121-27.
- Berg, B., Pettersson, G., Location and formation of cellulases in Trichoderma viride. J Appl Bacteriology, 1977; 42: 65-75.
- Berghem, L.E.R., Pettersson, L.G.,The mechanism of enzymatic cellulose degradation. Purification of cellulolytic enzymes from Trichoderma viride active on highly ordered cellulose. Eur J Biochem, 1973; 37: 21-30.
- Bhat, M., Bhat, S., “Cellulase degrading enzymes and their potential industrial applications,” Biotechnology Advances, 1997; 15(3-4): 583-620.
- Chen, P.J., Wei, T.C., Chang, Y.T., Lin, L.P., Purification and characterization of carboxymethyl cellulase from Sinorhizobium fredii. Bot Bull Acad Sin, 2004; 45: 111-18.
- Dariot, D.J., Simonetti, A., Plinho, F., Brandeli. A., Purification and characterization of extracellular b-glucosidase from Monascus auroureus. J Microbiol Biotechnol, 2008; 18: 933-41.
- Das, M., Banerjee, R., Bal. S., Multivariable parameter optimization for the endoglucanase production by Trichoderma reesei RUT C30 from Ocimum gratissimum seed. Braz Arch Biol Technol, 2008; 51: 35-41.
- Dutta, T., Sahoo, R., Sengupta, R., Ray, S.S., Bhattacharyee, A., Ghosh, S., Novel cellulases from an extremophilic filamentous fungus Penicillium Citrinum: production and characterization. J Ind Microbiol Biotechnol, 2008; 35: 275-82.
- Fawole, O. B., Odunfa, S.A., Some factors affecting production of pectic enzymes by Aspergillus niger. Int Biodeter Biodegr, 2003; 52: 223–227.
- Gao, J., Weng, H., Zhu, D., Yuan, M., Guan, F., Yu-Xi., Production and characterization of cellulolytic enzymes from the thermoacidophilic fungal Aspergillus terreus M11 under solid state cultivation of corn stover. Bioresour Technol, 2008; 99:7623-29.
- Ghorai, S., Chowdhary, S., Pal, S., Banik, S.P., Mukherjee, S.P., Khowala, S., Enhanced activity and stability of cellobiase (b-glucosidase: EC 3.2.1.21) produced in the presence of 2-deoxy-D-glucose from the fungus Termitomyces clypeatus. Carbohydrate Res, 2010; 345: 1015-22.
- Henrissat, B.,”A new cellulase family,” Molecular Microbiology, 1997; 23(4): 848-849.
- Jagtap, S., Rao, M., Purification and properties of a low molecular weight 1,4-bea-D-glucan glucohydrolase having one active site for carboxy methyl cellulase and xylan from an alkalothermophilic Thermomonospora sp. Biochem Biophys Res Commun, 2005; 329(1): 111-6.
- Krishna, C., Solid state fermentation system: an overview. Crit Rev Biotechnol, 2005; 25: 1-30.
- Lamed, R., Setter, E., Kenig, R., Biotechnol Bioeng Symp 1997; 13: 799-804.
- Lowry, O. H., Rosebrough, N.J., Farr, A.L., Randall, R.J., Protein measurement with folin-phenol reagent. J Biol Chem, 1951; 193: 265-75.
- Mandels, M., Andreotti, R.E., Roche, C., Measurement of sacharifying cellulose. Biotechnol Bioeng Symp, 1976; 6: 21-23.
- Mart-Yanov, V. A., Rabinovich, M.L. , Klesov, A.A., Myaghikh, I.V. , Gervert, M.V., Priki Biokhim Microbiol, 1983; 19: 513-20.
- Nagendran, S., Hallen-Adams, H.E., Paper, J.M., Aslam, N., Walton, J.D., Reduced genomic potential for secreted plant cell wall degrading enzymes in the ectomycorrhizal fungus Amanita bisporigera, based on the secretome of Trichoderma reesei. Fungal Gene Biol., 2009; 46: 427–35. doi: 10.1016/j.fgb.2009.02.001.
- Noronha, E. F., Kipnis, A., Kipinis, A.P.J., Ulhoa, C.J., Regulation of 36kDa 1-3-glucanse synthesis in Trichoderma harzianum. FEMS Microbial Lett, 2000; 188: 19-22.
- Ojumo, T. V., Solomon, B.O., Betiku, E., Loyokun, S.K., Amigun, B., Cellulase production by Aspergillus flavus L isolate NSPR 101 fermented in sawdust, bagasse and corncub. Afr J Biotechnol, 2003; 2: 150-52.
- Okada, G., Enzymatic studies on a cellulase system of Trichoderma viride: Purification and Properties of a less random type cellulase. J Biochem, 1976; 80: 913-22.
- Okada, G., Nisizawa, K., J Biochem, 1975; 78: 297-300.
- Perez, J., Munoz-Dorado,J., de-la-Rubia,T., Martinez, J., “Biodegradation and biological treatments of cellulose, hemicellulose and lignin: an overview,” International Microbiology, 2002; 5(2): 53-63.
- Pitson, S.M., Seviour, R.J., McDougall, M.B. Purification and characterization of an extracellular b-glucosidase from the filamentous fungus Acremonium persicinium and its probable role in b-glucan degradation. Enzyme Microbial Technol, 1997; 21: 182-90.
- Roberto, D.S., Lago, E.S., Merheb, C.W., Macchiore, M.M. , Park, Y.K., Gomes,E., Production of xylanase and CMCase on solid state fermentation in different residue by Thermoascus aurantiacus miehe. Braz J Microbiol, 2005; 36: 235-41.
- Rodriguez-couto, S., Sanroman, M.A., Application of solid state fermentation to ligninolytic enzyme production. Biochem Eng J, 2005; 22: 211-19.
- Roukas, T., Citric and gluconic acid production from fig by Aspergillus niger using solid- state fermentation. J Ind Microbiol Biotechnol, 2000; 25: 298-304.
- Roy, I., Sharma, A., Gupta, M.N., Recovery of biological activity in reversibly inactivated protein by three phase partitioning. Enzyme Microb Technol, 2005; 37: 113-20.
- Sakon, J., Irwin, D., Wilson, D.B., Karplus, P.A., “Structure and mechanism of endo/exocellulase E4 from Thermospora fusca,” Nature Structural Biology, 1997; 4(10): 810-818.
- Shanjing, Y., Yixin, G., Lihua,Y., Characterization of crude cellulase from Trichoderma reesei and purification of cellulase. Chinese J Chem Eng, 2002; 10(6): 723-25.
- Thongekkaew, J., Hiroko-Ikeda, H., Masaki, K., Iefuji,H., An acidic and thermostable carboxymmethylcellulase from the yeast Cryptococcus sp. S-2: purification and characterization and improvement if its recombinant enzyme production by high cell density fermentation of Pichia pastoris. Protein Expression Purif, 2008; 60(2): 140-6.
- Tomme, P., Warren, R.A., Gilkes, N.R., “Cellulose hydrolysis by bacteria and fungi,” Advances in Microbial Physiology, 1995; 37(1): 1-81.
- Toyama, N.,Ogawa,K., In: Ghose T K (Ed.), International Course on Biochemical Engineering Bioconversion,1977.
- Wei, D L, Kohtaro, K., Shoji, U., Hui, L .T., Purification and characterization of an extracellular b-glucosidase from the wood grown fungus Xylaria regalis. Curr Microbiol, 1996; 33: 297-301.
- Wood, T.M., McCrae,S.I., The purification and properties of the C1 component of Trichoderma koningii cellulase. Biochem J, 1972; 128: 1183-92.
- Xin, F., Geng,A., Horticultural waste as the substrate for cellulase and hemicellulase production by Trichoderma reesei under solid state fermentation. Appl Biochem Biotechnol, 2010; 162: 295-306.
- Yang, S., Jiang, Z., Yan, Q., Zhu,H., Characterization of a thermostable extracellular b-glucosidase with activities of exoglucanase and transglycosylation from Paecilomyces thermophila . J Agri Food Chem, 2008; 56: 602-08.
- Youssef, G.A., Physiological studies of cellulase complex enzymes of Aspergillus oryzae and characterization of carboxymethylcellulase. Afr J Microbiol Res, 2011; 5(11): 1311-21.
© The Author(s) 2017. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.


