ISSN: 0973-7510
E-ISSN: 2581-690X
Panton-Valentine Leukocidin (PVL) is a cytotoxin produced by Staphylococcus aureus that causes leukocyte destruction and tissue necrosis. Therefore, this study aimed to detect the rate, antimicrobial susceptibility, associated risk factors, and the phylogenetic relationship of PVL-positive S. aureus nasal carriers among patients and nurses. The research methods included the collection of 315 nasal specimens obtained from inpatients and nurses. The identification of S. aureus was confirmed by a coagulase test. The multiplex polymerase chain reaction technique was used to affirm PVL-positive S. aureus. The antibiotic sensitivity of S. aureus isolates was carried out using the disk diffusion method. The phylogenetic similarity of PVL-positive S. aureus was identified by pulsed-field gel electrophoresis. This study revealed that 160 out of 315 (50.8%) isolates were S. aureus. In addition, 7/160 (4.4%) had the lukS gene (six MSSA and one MRSA). The PVL-positive S. aureus isolates were 100% sensitive to gentamicin, linezolid, mupirocin, rifampin, trimethoprim-sulfamethoxazole, teicoplanin, tigecycline, and vancomycin. The risk factor analysis revealed that a longer hospital stay, nasogastric intubation, and runny nose were significant risk factors for patients to be PVL-positive S. aureus nasal carriers. A phylogenetic similarity analysis of PVL-positive isolates showed five models and they were distantly correlated. Therefore, the current study will provide knowledge to the hospital infectious control authority.
S. aureus, Nasal Carriers, PVL, Multiplex PCR, Antimicrobial Susceptibility, PFGE
Staphylococcus aureus (SA) is a nasal commensal that can be found in 30% of the population, one-third of which are persistently colonised.1 In particular, SA produces different virulence factors that facilitate nasal colonisation, as well as invade and evade the immune system. Panton-Valentine Leukocidin (PVL) is encoded by lukS and lukF-PV, which was confirmed to play a vital pathogenic role in SA by attacking monocytes and macrophages to create holes in the cell membrane and disrupts the permeability barrier of leukocytes and erythrocytes.2, 3 PVL can induce neutrophil lysis or apoptosis and lead to tissue necrosis.4 This process is followed by an outflux of cytokines, invigoration of intracellular proteases, initiation of apoptosis, and eventually, cell death.5 Moreover, PVL is also associated with skin and soft tissue infections, necrotising pneumonia, as well as osteomyelitis.6,7 Necrotising pneumonia rapidly destroys lung tissue and results in death in 75% of cases.7 Moreover, the PVL is more highly associated with methicillin-resistant SA (MRSA) compared to methicillin-susceptible SA (MSSA).8,9 Yoong and Torres reported that approximately 5% of SA clinical isolates possess PVL toxin.10 This study aims to detect the percentage of PVL-positive SA, antimicrobial susceptibility, related risk factors, and phylogenetic association of PVL-positive SA nasal carriers among both inpatients and nurses.
Study design and setting
This cross-sectional prospective study was performed from May 2017 to April 2018 at Hospital Sungai Buloh (HSB), a tertiary hospital in Selangor, Malaysia which has been established as a centre of excellence for infectious diseases. A total of 315 nasal swabs were collected from patients and nurses from three wards (medical, gynaecological, and surgical). Demographic and clinical data were collected from these records.
Bacterial isolates
The identification of SA isolates was performed using a Staphylase test kit (Oxoid, Basingstoke, England). Subsequently, a single colony was obtained, cultured onto blood agar plates, and incubated at 37°C overnight for further processing. All none SA isolates were discarded.
DNA extraction
Bacterial DNA was extracted using the boiling method. Pure SA colonies were mixed with 100 µL of sterile distilled water, placed in a 1.5 mL Eppendorf tube and incubated for 10 min at 99°C on a Microplate thermoshaker (QInstruments, Hamburg, Germany). The tube was then spun for 1 min at 1,500 g. Approximately 3 µL of the supernatant was collected to be added to the PCR mastermix.
Multiplex Polymerase Chain Reaction
The monoplex and multiplex PCR amplifications were performed using an Eppendorf Gradient Thermal Cycler. A final volume of 20 µL containing 10 μM dNTPs (0.8 μL), 25 mM MgCl2 (2.5 μl), 10X PCR buffer (2.0 μL), 5U Taq DNA polymerase (0.15 μL), 0.3 pmol of each primer (femA-F: 5’ GCAGCTTGCTTACTTACTG 3’; femA-R: 5’ TCCATCCATATTTTTAATGATG 3’; mecA-F; 5’ GATGGTATGTGGAAGTTAGA 3’; mecA-R: 5’ GAAATACTTAGT TCTTTAGCG 3’: lukS-F: 5’ GCAATGAGGTGGCCTTTC 3’ and lukS-R: 5’ TCATGA GTTTTCCAGTTCAC 3’) were used. The PCR cycling conditions consisted of: one cycle of initial denaturation at 94°C for 3 min, 30 cycles of denaturation at 94°C for 30 s, annealing for 30 s at 57.7°C, extension at 72°C for 1 min, and a final extension at 72°C for 5 min.
Antimicrobial Susceptibility
Antimicrobial susceptibility of PVL-positive SA isolates was performed using the disc diffusion technique with Mueller-Hinton agar as stated by CLSI guidelines.11 Bacterial broth for each isolate was created by emulsifying a few colonies from an overnight pure culture in Luria Bertani broth (Difco Laboratories, Le Pont de Claix, France) in Eppendorf tubes. Later, the turbidity was modified to the 0.5 McFarland standard. The bacterial broth was uniformly streaked on a Mueller Hinton agar surface and left for 3 min before applying antibiotics. The most common antimicrobial used for treatment of SA infections was selected, including ciprofloxacin (5 μg), fusidic acid (10 μg), gentamicin (10 μg), linezolid (30 μg), mupirocin (5 μg), oxacillin (1 μg), rifampin (5 μg), teicoplanin (30 μg), telithromycin (15 μg), tigecycline (15 μg), trimethoprim-sulfamethoxazole (25 μg), and vancomycin (30 μg). The plates were incubated at 37°C overnight, and the inhibition zones were measured. The results were expressed as stated by CLSI.11
Phylogenetic analysis of PVL isolates
Pulsed-field gel electrophoresis (PFGE) was performed according to the Centers for Disease Control and Prevention. DNA molecules was broken down into smaller pieces by SmaI restriction enzyme then loaded into 1% SeaKem Gold agarose gel at 15°C in TBE via a CHEF DR III. PFGE was carried out at 200 V using a switch on time of 5 s and switch off time of 40 s, the operating angle was 120°C, and the total running time was 21 h. Following the completion of electrophoresis, the agarose gel was stained with Gel Red (Biotium, Fremont, USA) for 15 min, and destained with distilled water for 40 min. The gel was then placed on a UV box and documented using Gel Doc 2000 (Bio-Rad Laboratories, California, USA). An analysis of the phylogenetic tree of the PVL-positive isolates was performed using Bionumerics software, version 7.5 (Applied Maths, Kortrijk, Belgium). Band styles of the PFGE pattern was interpreted according to the Dice coefficient and unweighted pair group matching analysis (UPGMA) set up based on the criteria reported by Tenover et al. (1995).12 Bacterial sequences with a coefficient of similarity ≥ 80% were counted as one pulsotype and specified alphabetically.
Statistics
The data were transferred to IBM SPSS Statistics for analysis. A P value < 0.05 was determined to be significant. Multiple logistic regression analysis was used to predict or explain the probability of PVL-positive SA nasal carriers with the associated risk factors of nasal colonisation.
S. aureus and the PVL encoding gene
Among the 315 specimens, 160 (50.8%) isolates were SA. The presence of the lukS gene for the PVL toxin was investigated, and lukS was detected in seven (4.4%) SA isolates by PCR. From the seven PVL-positive isolates, six were MSSA and one isolate was MRSA.
Antimicrobial susceptibility of PVL-positive isolates
Of the PVL-positive MSSA isolates (n = 6), all isolates were sensitive to gentamicin, linezolid, mupirocin, rifampin, trimethoprim-sulfamethoxazole, teicoplanin, tigecycline, and vancomycin. However, five isolates were sensitive to both ciprofloxacin and fusidic acid (83.3%), and four isolates were sensitive to telithromycin (66.7%). In this study, only one PVL-positive MRSA was detected, which was sensitive to all antibiotics except oxacillin and ciprofloxacin.
Risk factors for PVL-positive S. aureus nasal carriers
Patients with a nasogastric intubation or who presented with fever, runny nose, and stayed in the hospital for a long time had a higher risk of being PVL-positive SA nasal carriers (Table 1). However, Table 2 showed that the excess length of hospital stay was significantly (P = 0.004) higher among PVL-positive SA patients with mean (SD) 11 (± 4.08) compared to that of PVL-negative SA patients 4.23 (±4.51).
Table (1):
Multiple Logistic Regression Analysis of the Risk Factors of PVL-Positive SA Nasal Carriers.
| Variables | B | Wald | Adjusted OR | 95% CI | P value | |
|---|---|---|---|---|---|---|
| Lower | Upper | |||||
| Nose picking | 0.63 | 0.25 | 1.88 | 0.16 | 22.83 | 0.62 |
| Fever past 2 weeks | 1.80 | 2.03 | 14.40 | 1.69 | 123.05 | 0.01* |
| Antibiotic intake past 2 weeks | 0.69 | 0.70 | 0.50 | 0.10 | 2.53 | 0.4 |
| Nasogastric tube | 3.07 | 5.95 | 21.51 | 1.83 | 253.37 | 0.01* |
| Frequently sneeze | 0.21 | 0.05 | 0.81 | 0.14 | 4.68 | 0.82 |
| Nose itching | 2.33 | 7.31 | 10.25 | 1.90 | 55.42 | 0.01* |
| Runny nose | 2.99 | 6.31 | 19.78 | 1.93 | 202.99 | 0.01* |
| Snore at night | 0.34 | 0.15 | 1.41 | 0.24 | 8.21 | 0.7 |
| Difficult to breath | 0.77 | 0.95 | 0.46 | 0.46 | 10.26 | 0.33 |
| Sinusitis | 1.18 | 0.98 | 0.31 | 0.03 | 3.16 | 0.32 |
| Allergy | 0.58 | 0.27 | 1.79 | 0.19 | 16.58 | 0.61 |
*Significantly difference
Table (2):
Numerical Variables of PVL-Positive SA and PVL-Negative SA.
Variables |
PVL-Positive SA Mean (SD) |
PVL-Negative SA Mean (SD) |
Mean diff. (95% CI) |
t-statistic (df) |
P value |
|---|---|---|---|---|---|
Age |
53.7 (11.38) |
46.2 (18.09) |
7.4 (-6.7, 21.1) |
1.08 (158) |
0.28 |
Duration of Hospitalization |
11 (4.08) |
4.23 (4.51) |
6.7 (2.9, 10.5) |
4.271
(6.69) |
0.004* |
*Significantly difference
Phylogenetic analysis of PVL isolates
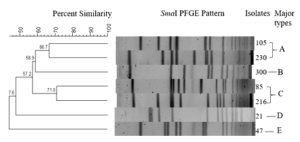
The phylogenetic relationship of all (n = 7) PVL-positive SA isolates was detected by PFGE (Figure 1). A percent similarity dendrogram with Dice coefficients PFGE data and a cut-off of 80% showed five patterns of isolates, and was designated as a pulsed-field type A to E (Figure 2). Since the percentage of similarity amongst the isolates was less than 80%, indicated that the isolates were distantly related.
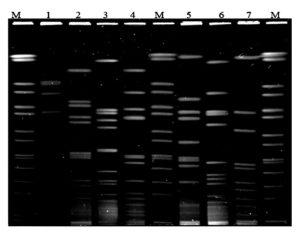
Figure 1. Pulsed-Field Gel Electrophoresis Patterns of sma1 Digested Genomics DNA of PVL-Positive Isolates from Patients in Hospital Sungai Buloh (Lane M, Salmonella braenderup as Standard Marker; Lane 1, PVL Isolate from Patient No. 21; Lane 2, PVL Isolate from Patient No. 47; Lane 3, PVL Isolate from Patient No. 85; Lane 4, PVL Isolate from Patient No. 105; Lane 5, PVL Isolate from Patient No. 171; Lane 6, PVL Isolate from Patient No. 216; Lane 7, PVL Isolate from Patient No. 230)
PVL-positive SA was confirmed by detecting the PVL encoding gene using molecular techniques. The current study showed seven PVL-positive SA isolates, which are six isolates from MSSA and one isolate from MRSA. All of the PVL-positive isolates were from the patient samples. Therefore, the rate of PVL-positive SA isolated in patients warded in HSB was 4.4%, which was similar to the previous studies, which showed 4.9% from Kuala Lumpur13 and 4.3% from Indonesia.14 In this study, more of the PVL-positive isolates were among MSSA than MRSA, which is in agreement with studies in Tehran and Kuwait.15,16 Unlike other studies, the PVL toxin gene was observed in the plurality of MRSA isolates.17,18 The rate of the PVL toxin gene among MSSA infections differs between countries and was reported to have a high rate (58%) in the African continent.19
In this study, PVL-positive SA were primarily MSSA, and susceptible to most antibiotics; however, PVL-positive MRSA was resistant to ciprofloxacin. Similar results have been reported by Bhatta et al. in Nepal, who reported that 77.2% of PVL-positive MRSA were resistant to ciprofloxacin.17 The PVL-positive SA isolates in the present study were sensitive to gentamicin, linezolid, mupirocin, rifampin, trimethoprim-sulfamethoxazole, teicoplanin, tigecycline, and vancomycin. These findings reflect the well-regulated use of these antibiotics in the hospitals.
Numerous variables were examined as a potential factors that were associated with PVL-positive SA nasal carriers. The present findings showed that a previous fever was closely related to PVL-positive nasal carriers. The results of this study were similar to earlier report by Gillet et al. who revealed that 76% of patients with PVL-positive SA nasal carrier had fever.20 This sign might give the impression that a person is being infected with PVL-positive SA.
Furthermore, this study demonstrates that the chances of being PVL-positive SA nasal carriers are 1.8 times higher among persons with a habit of nose picking; however, this was not significant (P = 0.62). Another study by Wertheim et al. stated that nose pickers had a significantly higher potential to carry SA than non-nose pickers.21 Transmission may occur when a person touches the surface or objects that have been contaminated by this bacterium and subsequently picking their nose. Such nose picking increases the likelihood of establishing bacterial colonisation in their nose due to SA being present on their finger. Occasionally, picking the nose may appear to be a more convenient means of cleaning out the nose instead of using tissue paper. Therefore, the person should wash his or her hands before and after picking their nose to prevent the spread of potential infectious organisms.
In addition, other factors that showed an increase risk for PVL-positive SA nasal carriers consisted of nose itching and a runny nose (P < 0.05). Contaminated hands represented the main vector for SA transmission from surfaces to the nose.21 The fingers are typically contaminated and easier to transmit bacteria to the nasal cavities. Similarly, where a person tends to wipe a runny nose presents the possibility of transmitting bacteria to the nose directly.
Another important cofounder was nasogastric intubation. In this study, a significant association (P = 0.01) was observed between PVL-positive nasal carriers and patients undergoing intubation compared to those who did not. This supports the findings of a former study in Malaysia by Al-Talib et al., which demonstrated that a nasogastric intubation is a risk factor for the acquisition of a nosocomial infection.22 A systemic review by Wu et al. listed nasogastric intubation as a risk factor for SA pneumonia,23 acting as a vehicle and assisting in the transmission of PVL-positive bacteria to the respiratory tract.
The current study revealed that the excess length of hospitalisation of 11 to 15 days was significantly higher (P = 0.004) among PVL-positive SA nasal carriers compared to PVL-negative SA individuals. Our results were in accordance with those of a previous study in Latvia, who reported that patients infected with PVL-positive SA required longer hospitalization than PVL-negative infections.24 These findings indicate that patients requiring a longer hospitalisation are generally those who have critical illness with low immunity. Thus, a longer hospitalisation will increase the risk of developing PVL nasal colonisation.
No significant difference (P > 0.05) was found between PVL nasal carriers and non-carriers regarding antibiotic intake during past two weeks (P = 0.40). Different results were reported by Bhutia and Singh in their study in India, which revealed that previous antibiotic intake had a significant effect on the chances of acquiring PVL-positive SA among their clinical isolates.25 In contrast, this study reported an odds ratio of 0.50, suggesting that an individual who takes an antibiotic is 50% less likely to be a PVL nasal carrier. Unfortunately, the type of antibiotic that has been taken by patients was unknown to be related to the susceptibility or resistance rate for PVL nasal carrier.
In the present study, the phylogenetic relationship between seven PVL-positive SA isolates was investigated. The phylogenetic relationships were examined using PFGE. PFGE is an extremely distinctive technique for SA patterns since it is relies on fragmentation of the bacterial chromosome using SmaI enzyme and detaching of the digestant DNA fragments on agarose gel using alternating electrical pulses. Five distinctive combined genotypes were identified among the seven SA isolates. Since the percentage of similarity amongst the isolates was less than 80% (counted with Dice coefficients from the PFGE data with a level of 80%), the PVL isolates were distantly related. Moreover, the isolates were not collected during outbreak situations. An earlier study conducted by Vivoni et al.26 in Brazil reported that a variety of PFGE patterns for MSSA, since their clinical specimens were obtained during an endemic and did not exhibit any clear dominant genotypes.26 Therefore, the authors hypothesized that Tenover’s standards were not readily appropriate since it was designed for outbreak circumstances, in which the PFGE band patterns are matched to that of an outbreak strain. Furthermore, isolates from Patterns A, C, D, and E differed because they belonged to different wards, and the source of transmission was difficult to identify due to the small number of PVL isolates.
The findings of this study suggested that the percentage rate of the PVL-positive SA was fairly low among inpatients and staff nurses in HSB. Moreover, none of the nurses were carriers of PVL positive SA. Patients with history of previous fever, nasogastric intubation, and itchy nose had higher chance of being PVL-positive SA nasal carriers. The possibility of PVL-positive SA cross transmission between different patients in HSB has been ruled out. The PVL toxin gene may represent a potential marker to reveal nosocomial infections among various patients who carry the same characteristic pathogen.
ACKNOWLEDGMENTS
The authors would like to thank HSB for their Endless Cooperation and Faculty of Medicine, IMMB Research Institute-UiTM, Malaysia.
CONFLICT OF INTEREST
The authors declared there is no conflict of interest.
AUTHORS’ CONTRIBUTION
HTB and ZMZ supervised the entire work. SS performed all laboratory work. HTB principal investigator of the grant. SS wrote the initial draft. HTB and ZMZ edited and reviewed the manuscript. HTB approved the manuscript for publication.
FUNDING
This study was funded by Internal Grant from Universiti Teknologi MARA 600-IRMI/GIP 5/3 (034/2019).
DATA AVAILABILITY
All datasets generated and analysed during this research are included in the manuscript.
ETHICS STATEMENT
This study was approved by the Institutional Ethics Committee, Universiti Teknologi MARA 600-RMI (5/1/6) and Medical Research Ethics Committee (MREC), Ministry of Health Malaysia NMRR-15-2091-27031 (IIR).
- Sakr A, Bregeon F, Mege JL, Rolain JM, Blin O. Staphylococcus aureus Nasal Colonization: An Update on Mechanisms, Epidemiology, Risk Factors, and Subsequent Infections. Front Microbiol. 2018;9:2419.
Crossref - Al-Talib H, Hasan H, Yean CY, Al-Ashwal SM, Ravichandran M. Fatal necrotizing pneumonia caused by Panton-Valentine Leukocidin-producing hospital-acquired Staphylococcus aureus: a case report. Jpn J Infect Dis. 2011;64(1):58-60.
Crossref - Norhafez S, Al-Talib H, Adnan A. Detection of Toxins and Antibiotic Resistance Genes Profile among Methicillin-Resistant Staphylococcus aureus (MRSA) Isolates and their types of Infection in a Tertiary Hospital in Malaysia. Int J Med Res Health Sciences. 2021;10(2):151-159. https://www.ijmrhs.com/medical-research/detection-of-toxins-and-antibiotic-resistance-genes-profile-among-methicillinresistant-staphylococcus-aureus-mrsa-isolat.pdf
- Tristan A, Bes M, Meugnier H, et al. Global distribution of Panton-Valentine Leukocidin-positive methicillin-resistant Staphylococcus aureus, 2006. Emerg Infect Dis J. 2007;13(4):594-600.
Crossref - Tomita T, Kamio Y. Molecular biology of the pore-forming cytolysins from Staphylococcus aureus, alpha- and gamma-hemolysins and Leukocidin. Biosci Biotechnol Biochem. 1997;61(4):565-752.
Crossref - Galia L, Ligozzi M, Bertoncelli A, Mazzariol A. Real-time PCR assay for detection of Staphylococcus aureus, Panton-Valentine Leucocidin and Methicillin Resistance directly from clinical samples. AIMS Microbiol. 2019;5(2):138-146.
Crossref - Gillet Y, Issartel B, Vanhems P, et al. Association between Staphylococcus aureus strains carrying gene for Panton-Valentine Leukocidin and highly lethal necrotising pneumonia in young immunocompetent patients. Lancet. 2002;359(9308):753-759.
Crossref - Darboe S, Dobreniecki S, Jarju S, et al. Prevalence of Panton-Valentine Leukocidin (PVL) and Antimicrobial Resistance in Community-Acquired Clinical Staphylococcus aureus in an Urban Gambian Hospital: A 11-Year Period Retrospective Pilot Study. Front Cell Infect Microbiol. 2019;9:170.
Crossref - Goudarzi H, Goudarzi M, Sabzehali F, Fazeli M, Salimi Chirani A. Genetic analysis of methicillin-susceptible Staphylococcus aureus clinical isolates: High prevalence of multidrug-resistant ST239 with strong biofilm-production ability. J Clin Lab Anal. 2020;34(11):e23494.
Crossref - Yoong P, Torres VJ. The effects of Staphylococcus aureus leukotoxins on the host: cell lysis and beyond. Curr Opin Microbiol. 2013;16(1):63-69.
Crossref - Clinical Laboratory Standards Institute. 2014. Performance Standards for Antimicrobial Susceptibility Testing: Eighteenth Informational Supplement. Clinical Laboratory Standards Institute; Wayne, PA, USA. 2014. CLSI Document M100-S18.
- Tenover FC, Arbeit RD, Goering RV, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33(9):2233-2239.
Crossref - Ghaznavi-Rad E, Nor Shamsudin M, Sekawi Z, et al. Predominance and emergence of clones of hospital-acquired methicillin-resistant Staphylococcus aureus in Malaysia. J Clin Microbiol. 2010;48(3):867-872.
Crossref - Santosaningsih D, Santoso S, Budayanti NS, et al. Epidemiology of Staphylococcus aureus harboring the mecA or Panton-Valentine Leukocidin genes in hospitals in Java and Bali, Indonesia. Am J Trop Med Hyg. 2014;90(4):728-734.
Crossref - Tajik S, Najar-Peerayeh S, Bakhshi B. Hospital clones of Panton-Valentine Leukocidin-positive and methicillin-resistant Staphylococcus aureus circulating in the Tehran community. J Glob Antimicrob Resist. 2020;22:177-181.
Crossref - Vali L, Dashti AA, Mathew F, Udo EE. Characterization of Heterogeneous MRSA and MSSA with Reduced Susceptibility to Chlorhexidine in Kuwaiti Hospitals. Front Microbiol. 2017;8:1359.
Crossref - Bhatta DR, Cavaco LM, Nath G, et al. Association of Panton Valentine Leukocidin (PVL) genes with methicillin resistant Staphylococcus aureus (MRSA) in Western Nepal: a matter of concern for community infections (a hospital based prospective study). BMC Infect Dis. 2016;16:199.
Crossref - Kaur H, Purwar S, Saini A, et al. Status of Methicillin Resistant Staphylococcus aureus Infections and Evaluation of PVL Producing Strains in Belgaum, South India. Journal of Krishna Institute of Medical Sciences University. 2012;1(2):43-51. http://www.jkimsu.com/jkimsu-vol1no2/jkimsu-vol1no2-OA-1-43-51.pdf
- Breurec S, Fall C, Pouillot R, et al. Epidemiology of methicillin-susceptible Staphylococcus aureus lineages in five major African towns: high prevalence of Panton-Valentine Leukocidin genes. Clin Microbiol Infect. 2011;17(4):633-639.
Crossref - Gillet Y, Vanhems P, Lina G, et al. Factors predicting mortality in necrotizing community-acquired pneumonia caused by Staphylococcus aureus containing Panton-Valentine Leukocidin. Clin Infect Dis. 2007;45(3):315-321.
Crossref - Wertheim HFL, van Kleef M, Vos MC, Ott A, Verbrugh HA, Fokkens W. Nose picking and nasal carriage of Staphylococcus aureus. Infect Control Hosp Epidemiol. 2006;27(8):863-867.
Crossref - Al-Talib H, Yean CY, Hasan H, Nik Zuraina NM, Ravichandran M. Methicillin-resistant Staphylococcus aureus nasal carriage among patients and healthcare workers in a hospital in Kelantan, Malaysia. Pol J Microbiol. 2013;62(1):109-112.
Crossref - Wu D, Wu C, Zhang S, Zhong Y. Risk Factors of Ventilator-Associated Pneumonia in Critically III Patients. Front Pharmacol. 2019;10:482.
Crossref - Cupane L, Pugacova N, Berzina D, Cauce V, Gardovska D, Miklasevics E. Patients with Panton-Valentine Leukocidin positive Staphylococcus aureus infections run an increased risk of longer hospitalisation. Int J Mol Epidemiol Genet. 2012;3(1):48-55. https://pubmed.ncbi.nlm.nih.gov/22493751/
- Bhutia KO, Singh TS. The Prevalence and the Risk Factors Which are Associated with Staphylococcus aureus and Methicillin-Resistant S. aureus Which Harboured the Panton- Valentine Leukocidin Gene in Sikkim. J Clin Diagn Res. 2012;6(3):393-399. https://www.jcdr.net/articles/PDF/1986/3981_E(C)_F(T)_PF(V)_PFA(A)_P(_)[1].pdf
- Vivoni AM, Diep BA, de Gouveia Magalhaes AC, et al. Clonal composition of Staphylococcus aureus isolates at a Brazilian university hospital: identification of international circulating lineages. J Clin Microbiol. 2006;44(5):1686-1691.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.