ISSN: 0973-7510
E-ISSN: 2581-690X
Panchagavya has traditionally been used in Indian Ayurvedic practices because of its pro-agricultural and medicinal properties. This study presents the draft genome of a new Brevibacillus brevis S1-3 strain isolated from the fermented product Panchagavya. Through whole-genome sequencing, we determined that the genome of B. brevis S1-3 was 6,348,716 base pairs with a GC content of 54.3%. Genome assembly revealed the presence of 6107 protein-coding genes, 186 tRNA genes, and 13 rRNA genes. Genome annotation and analysis identified the genes involved in metabolism and other cellular processes. We also predicted the presence of several gene clusters associated with plant growth promotion, including indole acetic acid (IAA), gibberellic acid, ammonia, and nitrogen. Our study also revealed the genes responsible for the production of secondary metabolites that displayed a significant correlation with antimicrobial activity. Our results provide new insights into the genomic basis of the plant growth-promoting abilities of B. brevis and pave the way for further research in this field.
Brevibacillus brevis, Draft Genome Sequencing, Panchagavya, Plant Growth Promotion, Secondary Metabolite, Ayurvedic Practices
Brevibacillus brevis is a gram-positive, motile, rod-shaped, aerobic spore-forming bacterium known to be present in various environmental conditions, including soil and the animal guts.1,2 This bacterium has been shown to possess antimicrobial activity against soil-borne pathogens such as Phytophthora nicotianae and Ralstonia solanacearum,3,4 making it a potential control agent against plant pathogens. Additionally, B. brevis produces a variety of secondary metabolites, such as tyrocidine, grastin, and adenine, which are responsible for its antimicrobial activity.5-7 Brevibacillus brevis has also been studied to identify its role and interaction with plants and has been found to confer disease resistance against fungal agents in plants like tomatoes,8 grapes,9 pigeon pea,10 tea,11 etc. Furthermore, B. brevis has been identified as a plant-growth-promoting rhizobacterium (PGPR),12-14 which can act as a biofertilizer, increasing crop yield and soil fertility, while reducing the need for chemical fertilizers.15
Several studies have reported draft genome sequences of several strains of B. brevis, including NBRC 100599,16 B. brevis X23,14 and B. brevis strain FJAT-0809-GLX.13 These genome sequences typically range in size from 6Mb and contain more than 5600 protein-coding genes. However, these previously published genomes are yet to undergo functional annotation to identify the genes responsible for the biosynthesis of secondary metabolites or plant growth regulators.
In the field of plant-microbe interactions, biocontrol is a dynamic strategy that uses beneficial microbes to control plant pathogens. The biocontrol arsenal includes systemic resistance, antimicrobial compounds, competitive exclusion, and nutrient enhancement.17 The success of biocontrol depends on factors such as compatibility, adaptability, persistence, and specificity, which collectively determine its effectiveness. Integrating these methods with other pest management approaches is essential for sustainable agriculture.18 However, achieving a delicate balance between inducing resistance without harmful effects and addressing practical application challenges remains a complex task.
Recent advancements highlight the crucial roles of plant-associated microorganisms in maintaining plant health and ecological balance. Utilizing beneficial microbes is a promising approach for disease mitigation and improved crop yields.19 Genomic and proteomic analyses of microbial genomes provide insights into the molecular intricacies of these interactions, which are critical for refining control measures. While previous research focused on the rhizosphere, the phyllosphere, which includes aboveground plant parts, is less explored.20
B. brevis is recognized as a noteworthy inhabitant of the rhizosphere, showcasing remarkable biocontrol capabilities through its interactions with plants. In this study, we isolated a new strain of Brevibacillus brevis S1-3 from Panchagavya, a fermented product traditionally used in Indian Ayurvedic practices that is composed of five cow products, including clarified butter, curd, milk, urine, and fermenting dung.21,22 Through genome sequencing and functional annotation, we characterized the genome of B. brevis S1-3, providing new insights into the genomic basis of the biosynthesis of secondary metabolites and plant growth regulators in this strain.
Isolation and Molecular Identification
The Panchagavya used in this study were obtained from a commercial market in Chennai, India. After serial dilutions, the bacterial species present in Panchagavya were grown in Luria-Bertani medium at 37°C. Distinct colonies were then selected and cultured separately before storage at -20°C. Genomic DNA was extracted from the selected bacteria using the QIAamp DNA Microbiome Kit (Qiagen India Pvt. Ltd., India), according to the manufacturer’s instructions. The quality and quantity of the extracted bacterial genomic DNA were analyzed using agarose gel electrophoresis and Nanodrop (Tecan-Infinite 200 PRO, Switzerland). PCR was performed using the 16s rDNA universal primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-TACGGTTACCTTGTTACGACTT-3′).23,24 The amplified PCR product was purified (using a Qiagen PCR product purification kit) and sequenced using the Sanger DNA sequencing method (Applied Biosystems Genetic Analyzer, Saint Aubin, France). The resulting 16s rDNA sequences were compared to those in the NCBI database using the Basic Local Alignment Search Tool (BLAST). The bacterial species were identified based on sequence similarity, and a phylogenetic tree was constructed using the MEGAX software. Evolutionary distances were inferred using the neighbour-joining method.25-27
Genome Sequencing and Annotation
Paired-end sequencing libraries were prepared using a Nextera XT DNA Library Preparation Kit (Illumina). The final library was analyzed using a Bioanalyzer 2100 (Agilent Technologies, USA) with a high-sensitivity DNA kit according to the manufacturer’s instructions. The paired-end Illumina library was sequenced using 2 x 150 bp chemistry on a NextSeq-500 sequencer. Quality control of the raw reads was performed using FastQC v.0.11.5,28-30 and the low-quality reads were filtered. The Cutadapt tool was used to remove adapter regions from sequencing reads. High-quality reads obtained from Illumina NextSeq-500 were assembled into scaffolds using SPAdes (version 3.7.1) with default parameters.31-33 The quality of the assembled genome was analyzed using QUAST.
Genome assembly was annotated using Prokka v.2.1.1 and Rapid Annotation using Subsystems Technology (RAST) server v.2.0. Secondary metabolite gene clusters were identified using the antiSMASH version 5. The various biological features of the annotated genome were analyzed using RAST. Antimicrobial resistance genes and other protein functions were identified using PATRIC genome analysis server.34-36
Isolation of culture and phylogenetic analysis
We isolated various bacterial strains and evaluated their antimicrobial activity. Antibacterial activity was examined using broth microdilution assays against Streptococcus aureus (NCBI_CP00253), E. coli (NCBI_U00096), and Vibrio cholerae (NCBI_CP043554). One of the bacterial isolates that exhibited antimicrobial activity was selected for this study.
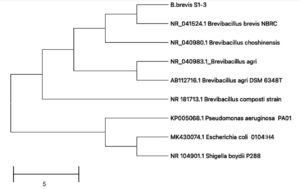
We isolated several bacterial strains from Panchagavya and assessed their antimicrobial potential. The antibacterial activity of these strains was evaluated using broth microdilution assays against three target pathogens: Streptococcus aureus, E. coli, and Vibrio cholerae (data not shown). One strain demonstrated notable antimicrobial activity among the bacterial isolates tested, prompting its selection for further investigation. The selected bacterial isolate was identified by 16s rDNA sequencing; and showed high similarity to Brevibacillus brevis (NR_041524). The 16s rDNA gene sequence of Brevibacillus brevis S1-3 was used to construct a phylogenetic tree (Figure 1), which revealed that the isolate was closely related to B. brevis NBRC and B. choshinensis with 99.2% and 98.38% sequence similarity, respectively. Other closely related species included B. agri and B. agri DSM 6348T, with 97.5% and 97.3% sequence similarity, respectively. The bacterial isolate identified in this study was named Brevibacillus brevis S1-3. The efficiency of Brevibacillus brevis as a plant growth-promoting rhizobacterium (PGPR) has been determined through studies evaluating its application in fostering plant growth.2 Through the examination of several plant growth-promoting (PGP) features, such as ammonia synthesis, and the generation of phytohormones, such as indole-3-acetic acid (IAA), Brevibacillus brevis’ efficiency in promoting plant growth, evaluations of seed germination, and several plant growth metrics have also been made.37 Bacillus brevis has been found to provide a multi-pronged defense against fungal and microbial pathogens by means of extracellular secretion of gramicidin S, gramicidin A, and a biosurfactant, thereby functioning as a biological control agent and aiding plant growth, apart from the production of PGPs.38
Figure 1. Phylogenetic analysis of 16S rDNA sequence of Brevibacillus brevis S1-3 strain using neighbor-joining method. Pseudomonas aeruginosa was used as an outgroup
Whole Genome Sequencing of B. brevis S1-3
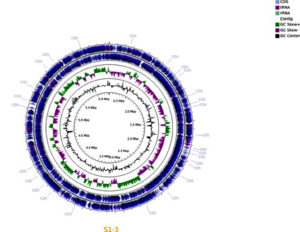
Genome sequencing of B. brevis S1-3 was performed using the Illumina NextSeq-500 platform. A total of 1,602,833 paired-end reads of 101bp were generated, with an average GC content of 54.3% (Table 1). These reads were assembled using SPAdes software, resulting in a draft genome of 5,845,263 bp in size, comprising 187 contigs (N50 – 88,031 bp) and 20 scaffolds (N50 – 678,417 bp). The genome contained 6,107 protein-coding sequences (CDS), 186 tRNA genes, and 13 rRNA operons (16S-23S-5S rRNA)
(Figure 2). Genome annotation was performed using the Prokka and RAST servers, which revealed that out of the total of 2,616 proteins, 2,492 were annotated as ‘hypothetical’ while the remaining proteins had non-hypothetical functions. The annotation included 5,259 proteins with functional assignments, including 1,592 proteins with Enzyme Commission numbers, 1,355 with Gene Ontology (GO) assignments, and 1,201 proteins mapped to KEGG pathways. The quality of the genome assembly was evaluated using QUAST and showed that the genome assembly of B. brevis S1-3 was of high quality.
Table (1):
General genome features of Brevibacilus brevis S1-3 strain plant growth promoting bacteria isolated from Panchagavya.
Features |
S1-3 chromosome |
|---|---|
Genome size |
6,348,716 |
G + C (%) |
55.2 |
Predicted CDS |
5800 |
rRNAs |
13 |
tRNAs |
186 |
G+C (%): guanine and cytosine content; CDS: protein-coding genes; rRNAs: ribosomal RNA; tRNAs: transfer RNA
Figure 2. The chromosome organization of Brevibacillus brevis S1-3, a plant growth-promoting bacteria isolated from Panchagavya. Circularized DNA plotter diagram of the chromosome of B. brevis, oriented from the origin; the outer light blue circle designates the genome base positions, and the outer blue circles depict predicted 5800 CDSs on both forward and reverse strands. The purple and green combination circle states important chromosomal core structures with DNA elements like tRNA, GC skew+, GC skew-, and rRNA contig. The inner black circle denotes GC content.
Genome annotation
Genome annotation of B. brevis S1-3 assigned many genes to cellular processes related to metabolism, such as membrane transport, dormancy and sporulation, cellular signalling and regulation, cell wall synthesis, and capsule formation. Additionally, many genes were correlated with biosynthesis of a diverse group of macromolecules, such as amino acids, carbohydrates, cofactors, vitamins, prosthetic groups, and pigments (Table 2). A similar study conducted on Brevibacillus brevis LABIM17 proved its antimicrobial property against plant pathogens by brevis through the production of octapeptin and, auranticin.39
Table (2):
Annotation of genes involved in metabolism and other cellular processes of Brevibacillus brevis S1-3 plant growth-promoting bacteria isolated from Panchagavya
| Genes function | Compounds | No. of genes |
|---|---|---|
| Genes related to metabolism | Fatty acids, lipids and isoprenoids | 215 |
| Amino acids and derivatives | 625 | |
| Sulphur | 57 | |
| Carbohydrates | 560 | |
| Cofactors, vitamins, prosthetic groups and pigments | 381 | |
| Aromatic compounds | 45 | |
| DNA | 140 | |
| Phosphorous | 85 | |
| Iron | 29 | |
| Secondary metabolism | 8 | |
| Nitrogen | 16 | |
| Nucleosides and nucleotides | 163 | |
| Potassium | 13 | |
| RNA | 207 | |
| Genes related to cellular processes | Cell division and cellular cycle | 56 |
| Dormancy and sporulation | 141 | |
| Cellular wall and capsule formation | 145 | |
| Photosynthesis | 0 | |
| Miscellaneous | 67 | |
| Motility and chemotaxis | 118 | |
| Regulation and cell signalling | 115 | |
| Phages, prophages, transposable elements and plasmids | 14 | |
| Respiration | 109 | |
| Response to stress | 124 | |
| Membrane transport | 226 | |
| Virulence, disease and defence | 128 |
Identification of genes involved in plant growth promotion and secondary metabolite biosynthesis
B. brevis also exhibits PGP traits at high temperatures, making it a valuable inoculant for cotton crops. Previous studies have reported that B. brevis enhances plant growth by increasing the expression of plant growth promoters such as IAA, ammonia, siderophores, cytokinins, and GA3.2,40,41 Analysis of B. brevis S1-3 revealed that the genome contains many genes involved in the biosynthesis of plant growth promoters (PGP) (Table 3). The presence of five structural genes, trpE, trpD, trpC, trpB, and trpA in B. brevis S1-3 predicted the indole acetic acid production through the tryptophan pathway.42 The amoA and amoCAB code ammonia monooxygenase, which is essential for ammonia production. nifD, nifK, and nifH are responsible for metabolism involved in nitrogen fixation. entA, entB, and entC encode 2,3-dihydro-2,3-dihydroxybenzoate synthetase, which is essential for siderophore production.43 Cytokinin production was predicted based on the presence of Tzs genes, which encode cytochrome P450 monooxygenase, the key enzyme for cytokinin production.44 ggs1 and ggs2 initiate the GGDP pathway for primary metabolism of gibberellic acid.45 The presence of these genes in B. brevis S1-3 suggests that this strain has potential applications in agriculture as a biofertilizer and for controlling plant pathogens.
Table (3):
Plant growth promotor (PGP) gene cluster identified in B. brevis S1-3 strain
Plant growth promotor |
Genes |
|---|---|
IAA (Indole Acetic Acid) |
Iaam, Iac, IaaH, IaaL, trpE(G), ipdC |
Ammonia and Nitrogen |
amoA, amoCAB, nifD, nifK, nifH |
Siderophore |
Sid, agbB, entB, entC, entA |
Cytokines |
Tzs, TLRs, PDGFA, PDGFB, PDGFC, PDGFD |
GA3 |
P450-3, P450-4, NPB20, ggs1, ggs2 |
The gene clusters involved in the biosynthesis of secondary metabolites in B. brevis S1-3 were identified using the antiSMASH 5.1.2 software (Table 4). This analysis revealed 97 genes associated with antibiotic resistance, 47 genes related to drug targets, 79 transporter genes, and 96 virulence factor genes. The genes were classified based on their antimicrobial resistance mechanisms, as determined by various antimicrobial resistance databases.46-49 This study provides a comprehensive understanding of the genomic basis for the plant growth-promoting and secondary metabolite biosynthetic abilities of B. brevis S1-3 and, provides a foundation for future research in this area.
Table (4):
Antimicrobial Resistance Genes from Brevibacillus brevis S1-3
AMR Mechanism |
Genes |
|---|---|
Antibiotic inactivation enzyme |
ANT(6)-I, FosB, PDC family |
Antibiotic target in susceptible species |
Alr, Ddl, dxr, EF-G, EF-Tu, folA, Dfr, folP, gyrA, gyrB, inhA, fabI, Iso-tRNA, kasA, MurA, rho, rpoB, rpoC, S10p, S12p |
Antibiotic target modifying enzyme |
Cfr |
Efflux pump conferring antibiotic resistance |
EmrAB-OMF, EmrAB-TolC, FexA family, MdtABC-OMF, MdtABC-TolC, MexAB-OprM, MexCD-OprJ, MexCD-OprJ system, MexEF-OprN, MexHI-OpmD, MexHI-OpmD system, MexJK-OprM/OpmH, MexVW-OprM, MexXY-OMP, YkkCD |
Gene conferring resistance via absence |
gidB |
Protein altering cell wall charge conferring antibiotic resistance |
GdpD, PgsA |
Protein modulating permeability to antibiotic |
OccD4/OpdT, OccD6/OprQ, OccK8/OprE, OprD family |
Regulator modulating expression of antibiotic resistance genes |
LiaF, LiaR, LiaS |
This study isolated and characterized a new strain of Brevibacillus brevis, designated as S1-3, from Panchagavya. The 16s rDNA sequencing and phylogenetic analysis revealed that the isolate was closely related to B. choshinensis and B. agri 5-2. Genome sequencing of B. brevis S1-3 revealed that the genome is of high quality and contains a wide range of genes involved in various cellular processes, including metabolism, cell wall synthesis, and capsule formation. In addition, the genome contains many genes involved in the biosynthesis of plant growth promoters and secondary metabolites. The presence of genes involved in the biosynthesis of indole acetic acid, ammonia, nitrogen fixation, siderophores, cytokinins, and gibberellic acid suggests that this strain has potential applications as a biofertilizer and in controlling plant pathogens. Furthermore, identifying the genes involved in antibiotic resistance, drug targets, transport, and virulence factors may provide insights into the potential biotechnological applications of this strain. The results of this study expand our understanding of the genetic and functional diversity of B. brevis and provide a foundation for future research.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
PR and MRam conceptualized the idea. SS, AA and VS isolated and sequenced the genome of Brevibacillus brevis S1-3. JJ and MRan performed Genome annotation. PR, VS and JJ wrote the manuscript. All authors read and approved the final manuscript for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- Thomas P. Isolation of an ethanol-tolerant endospore-forming Gram-negative Brevibacillus sp. as a covert contaminant in grape tissue cultures. J Appl Microbiol. 2006;101(4):764-774.
Crossref - Nehra V, Saharan BS, Choudhary M. Evaluation of Brevibacillus brevis as a potential plant growth promoting rhizobacteria for cotton (Gossypium hirsutum) crop. Springerplus. 2016;5(1):1-10.
Crossref - Jianmei C, Bo L, Zheng C, Huai S, Guohong L, Cibin G. Identification of ethylparaben as the antimicrobial substance produced by Brevibacillus brevis FJAT-0809-GLX. Microbiol Res. 2015;172:48-56.
Crossref - Che J, Liu B, Ruan C, Tang J, Huang D. Biocontrol of Lasiodiplodia theobromae, which causes black spot disease of harvested wax apple fruit, using a strain of Brevibacillus brevis FJAT-0809-GLX. Crop Protection. 2015;67:178-183.
Crossref - Westman EL, Yan M, Waglechner N, Koteva K, Wright GD. Self resistance to the atypical cationic antimicrobial peptide edeine of Brevibacillus brevis Vm4 by the N-acetyltransferase EdeQ. Chem Biol. 2013;20(8):983-990.
Crossref - Martens T, Gram L, Grossart HP, et al. Bacteria of the Roseobacter clade show potential for secondary metabolite production. Microb Ecol. 2007;54(1):31-42.
Crossref - Zhu Y, Chen K, Ding Y, et al. Metabolic and proteomic mechanism of benzo[a]pyrene degradation by Brevibacillus brevis. Ecotoxicol Environ Saf. 2019;172:1-10.
Crossref - Chandel S, Allan EJ, Woodward S. Biological Control of Fusarium oxysporum f.sp. lycopersici on Tomato by Brevibacillus brevis. J Phytopathol. 2010;158(7-8):470-478.
Crossref - Che J, Lai C, Lai G, Chen Q, Liu G, Liu B. Effects of a Mixture of Brevibacillus brevis with Other Bacillus sp. Strains against Gray Mold and on Enzyme Activities of Grape. Agronomy. 2023;13(7):1724.
Crossref - Bapat S, Shah AK. Biological control of fusarial wilt of pigeon pea by Bacillus brevis. Can J Microbiol. 2000;46(2):125-132.
Crossref - Yang W, Yang H, Bao X, et al. Brevibacillus brevis HNCS-1: a biocontrol bacterium against tea plant diseases. Front Microbiol. 2023;14:1198747.
Crossref - Jin F, Ding Y, Ding W, Reddy MS, Fernando WGD, Du B. Genetic diversity and phylogeny of antagonistic bacteria against Phytophthora nicotianae isolated from tobacco rhizosphere. Int J Mol Sci. 2011;12(5):3055-3071.
Crossref - Che J, Liu B, Lin Y, Tang W, Tang J. Draft Genome Sequence of Biocontrol Bacterium Brevibacillus brevis Strain FJAT-0809-GLX. Genome Announc. 2013;1(2):160-173.
Crossref - Chen W, Yunsheng W, Dingjun L, Lin L, Qiming X, Qingming Z. Draft Genome Sequence of Brevibacillus brevis Strain X23, a Biocontrol Agent against Bacterial Wilt. J Bacteriol. 2012;194(23):6634-6635.
Crossref - Prasad R, Kumar M, Varma A. Role of PGPR in Soil Fertility and Plant Health. Soil Biology, 2015:247-260.
Crossref - Panda AK, Bisht SS, DeMondal S, Senthil Kumar N, Gurusubramanian G, Panigrahi AK. Brevibacillus as a biological tool: a short review. Antonie Van Leeuwenhoek. 2014;105(4):623-639.
Crossref - Ongena M, Jacques P. Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends Microbiol. 2008;16(3):115-125.
Crossref - Heydari A, Pessarakli M. A review on biological control of fungal plant pathogens using microbial antagonists. J Biol Sci. 2010;10(4):273-290.
Crossref - Montesinos E. Plant-associated microorganisms: A view from the scope of microbiology. Int Microbiol. 2003;6(4):221-223.
Crossref - Vorholt JA. Microbial life in the phyllosphere. Nat Rev Microbiol. 2012;10(12):828-840.
Crossref - Somasundaram E, Amanullah MM, Vaiyapuri K, Thirukkumaran K, Sathyamoorthi K. Influence of organic sources of nutrients on the yield and economics of crops under maize based cropping system. Journal of Applied Sciences Research, 2007;3(12): 1774-1777.
- Tharmaraj K. A Critical Review on Panchagavya – A Boon Plant Growth. Int J Pharm Biol Arch. 2011.
- Jonasson J, Oloifsson M, Monstein HJ. Classification, identification and subtyping of bacteria based on pyrosequencing and signature matching of 16s rDNA fragments. 2002. APMIS. 2007;115(5):668-677.
Crossref - Amutha K, Kokila V. PCR Amplification, Sequencing of 16S rRNA Genes with Universal Primers and Phylogenetic Analysis of Pseudomonas aeruginosa. International Journal of Science and Research (IJSR), 2014; 3(8):257-261
- Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 2016;33(7):1870.
Crossref - Barazesh A, Sarkari B, Shahabi S, et al. Genetic Diversity of Echinococcus granulosus Isolated from Humans: A Comparative Study in Two Cystic Echinococcosis Endemic Areas, Turkey and Iran. Biomed Res Int. 2020.
Crossref - Mohamed MSM, El-Arabi NI, El-Hussein A, El-Maaty SA, Abdelhadi AA. Reduction of chromium-VI by chromium-resistant Escherichia coli FACU: a prospective bacterium for bioremediation. Folia Microbiol (Praha). 2020;65(4):687-696.
Crossref - Brown J, Pirrung M, Mccue LA. FQC Dashboard: integrates FastQC results into a web-based, interactive, and extensible FASTQ quality control tool. Bioinformatics. 2017;33(19):3137-3139.
Crossref - Miossec MJ, Valenzuela SL, Mendez KN, Castro-Nallar E. Computational Methods for Human Microbiome Analysis. Curr Protoc Microbiol. 2017;47(1).
Crossref - Bittencourt SA. FastQC: a quality control tool for high throughput sequence data – ScienceOpen. Accessed August 23, 2023. https://www.scienceopen.com/document?vid=de674375-ab83-4595-afa9-4c8aa9e4e736
- Bankevich A, Nurk S, Antipov D, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455-477.
Crossref - Nurk S, Bankevich A, Antipov D, et al. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J Comput Biol. 2013;20(10):714-737.
Crossref - Bean DC, Agarwal A, Cherian BP, Wareham DW. Hypermucoviscous polymyxin-resistant Klebsiella pneumoniae from Kolkata, India: Genomic and phenotypic analysis. J Glob Antimicrob Resist. 2019;17:1-2.
Crossref - Wattam AR, Davis JJ, Assaf R, et al. Improvements to PATRIC, the all-bacterial Bioinformatics Database and Analysis Resource Center. Nucleic Acids Res. 2017;45(D1):D535-D542.
Crossref - Davis JJ, Wattam AR, Aziz RK, et al. The PATRIC Bioinformatics Resource Center: expanding data and analysis capabilities. Nucleic Acids Res. 2020;48(D1):D606-D612.
Crossref - Gillespie JJ, Wattam AR, Cammer SA, et al. PATRIC: the Comprehensive Bacterial Bioinformatics Resource with a Focus on Human Pathogenic Species. Infect Immun. 2011;79(11):4286.
Crossref - Mowafy AM, Khalifa S, Elsayed A. Brevibacillus DesertYSK and Rhizobium MAP7 stimulate the growth and pigmentation of Lactuca sativa L. Journal of Genetic Engineering and Biotechnology. 2023;21(1).
Crossref - Seddon B, McHugh RC, Schmitt A. Brevibacillus brevis – a novel candidate biocontrol agent with broad-spectrum antifungal activity. The BCPC Conference: Pests and diseases, Volume 2 Proceedings of an international conference held at the Brighton Hilton Metropole Hotel, Brighton, UK. 2000. 2000:563-570.
- Larissa de MC, Manoel TG, Mirela M, et al. Draft Genome Sequence of Brevibacillus brevis LABIM17, a Biotechnologically Important Antimicrobial-Producing Bacterium. Microbiol Resour Announc. 2022;11(3):e0000622.
Crossref - Wang X, Zhang J, Wang X, et al. The Growth-Promoting Mechanism of Brevibacillus laterosporus AMCC100017 on Apple Rootstock Malus robusta. Hortic Plant J. 2022;8(1):22-34.
Crossref - Sheng M, Jia H, Zhang G, et al. Siderophore Production by Rhizosphere Biological Control Bacteria Brevibacillus brevis GZDF3 of Pinellia ternata and Its Antifungal Effects on Candida albicans. J Microbiol Biotechnol. 2020;30(5):689-699.
Crossref - Vivas A, Barea JM, Azcon R. Interactive effect of Brevibacillus brevis and Glomus mosseae, both isolated from Cd contaminated soil, on plant growth, physiological mycorrhizal fungal characteristics and soil enzymatic activities in Cd polluted soil. Environ Pollut. 2005;134(2):257-266.
Crossref - Vivas A, Biro B, Nemeth T, Barea JM, Azcon R. Nickel-tolerant Brevibacillus brevis and arbuscular mycorrhizal fungus can reduce metal acquisition and nickel toxicity effects in plant growing in nickel supplemented soil. Soil Biol Biochem. 2006;38(9):2694-2704.
Crossref - Sakakibara H. Cytokinin Biosynthesis and Regulation. Vitam Horm. 2005;72:271-287.
Crossref - Bomke C, Tudzynski B. Diversity, regulation, and evolution of the gibberellin biosynthetic pathway in fungi compared to plants and bacteria. Phytochemistry. 2009;70(15-16):1876-1893.
Crossref - Sayers S, Li L, Ong E, et al. Victors: a web-based knowledge base of virulence factors in human and animal pathogens. Nucleic Acids Res. 2019;47(D1):D693-D700.
Crossref - Saier MH, Tran CV, Barabote RD. TCDB: The Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Res. 2006;34(suppl_1):D181-D186.
Crossref - Wishart DS, Knox C, Guo AC, et al. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36(issue suppl-1):D901-D906.
Crossref - McArthur AG, Waglechner N, Nizam F, et al. The comprehensive antibiotic resistance database. Antimicrob Agents Chemother. 2013;57(7):3348-3357.
Crossref
© The Author(s) 2024. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.