ISSN: 0973-7510
E-ISSN: 2581-690X
Laccases are the widely distributed oxidative enzymes belong to the group of phenoloxidases. These are considered as model enzymes and widely distributed in nature. Bacterial laccases have extremophilic nature and several other unique properties that are not features of fungal laccase such as high-temperature stability and pH. The present study focused on the optimization of laccase production from Bacillus sp. AKRC01 under submerged fermentation state using agro-waste as the substrate. The optimized environmental conditions such as incubation period 96 hours, temperature 35°C and pH 8.0 were obtained maximum enzyme production 3.832 U/ml. In addition, optimized the carbon and nitrogen sources such as 2.0% glucose (4.967 U/mL) and 1.0% peptone (6.236 U/mL) resulted in maximum enzyme production with rice bran as an effective agro-waste substrate. Laccase enzyme was partially purified via ammonium sulfate precipitation method and chromatography using the sephadex column and determination of the molecular weight of 61 kDa compared with the protein ladder was done by sodium dodecyl sulfate polyacrylamide gel electrophoresis. The findings of this study demonstrated the ability of Bacillus sp. AKRC01 using low-cost agro-waste for laccase production. Laccases are important enzymes that have several industrial applications such as bioremediation, bio pulping and food industry.
Agro-waste, Bioremediation, Laccases, Optimization, Partial purification, SDS-PAGE.
The production of laccase enzyme is a recent advancement in the field of biotechnology. Due to extremophilic properties of laccase enzymes indicated their broad prospects in varied environmental conditions. The laccase enzyme plays a vital role in bioremediation of industrial waste. The laccase function for lignin and phenolic compounds degradation has been assessed due to their wide range of biotechnological applications such as degradation of dyes1,2, bioremediation of toxic chemical waste1,3, wastewater and soil treatment, and biosensors development4,5. Due to the oxidation capacity of phenols, polyphenols and high specificity of the substrate, laccase has gained great concern from researchers over the past decades. Laccases are commonly distributed in the large blue copper-containing protein in higher crops (turnips, cabbages, beets, tomato, asparagus, pears, apples, and other vegetables) as a broad spectrum, fungi (Ascomyceteous, Deuteromyceteous, and Basidiomyceteous) and bacteria (Azospirillum lipoferum, P. Putida, P. desmolyticum NCIM 2112, Bacillus sp. HR03, B. halodurans, B. pumilus, etc)6,7. The bacterial laccase is mostly highly thermotolerant and maintains its activity in acidic, alkaline and natural conditions8, whereas, production of fungal laccase generally decreased rapidly at elevated temperature and pH. Because of this, bacteria can tolerate a wide range of abiotic factors as compared to fungi. In addition, bacterial laccases have some additional advantages due to their cost-effective use in industrial applications, including large substrate specificity, short term enzyme production and simplicity of cloning and expressing with suitable manipulation in the host. Laccase production from agro-waste provides a great opportunity to handle agro-waste together with enzyme production for multiple industrial applications and minimize the environmental pollutions. The generation of agro-waste from nature and various agro-based industries indicated the magnitude of problems including pulp and paper mill, sugarcane molasses-based distilleries, agricultural waste, and food industry. In addition, these agro-waste substrates also provide a favorable condition and habitat for the large amount of lignocellulytic enzyme secretion9-12. Moreover, several other factors, i.e. media composition, time, pH and the varying concentrations of the source of carbon and nitrogen, influence laccase production13,14. Considering the importance of laccase in pollution degradation and that the bacterial laccase toolbox from different subfamilies is the need of time to bioprospecting new bacterial strain for laccase production.
The present work was evaluated the utilization of various kinds of agro wastes such as rice bran, wheat bran, sawdust, banana peel, orange peel, potato peel, pea peel and sugarcane bagasse to increase laccase production from Bacillus sp. AKRC01. This process makes use of another material that requires less energy, reduces the pollutants in environmental, as cost-effective and ecofriendly. The best-suited agro-waste was optimized by using the statistical tool response surface methodology (RSM)15-17. RSM is a set of valuable statistical and mathematical methods for designing laccase production processes, enhancing and optimizing variables18. Central composite design (CCD) is used for the optimization of laccase production19-22 in submerged fermentation state (SmF). Partial purification of the laccase enzyme was carried out using ammonium sulfate precipitation method23, sephadex column (G-100) chromatography and SDS-PAGE molecular weight determination. In addition, laccase characterization was also performed to explore their potential for wide biotechnology applications.
Isolation and screening of laccase producing bacterial strain
Paper mill sludge sample collected from the M/s Star Paper Mill in Saharanpur, UP India, they are located at Latitude: 29° 56’ 10.89” N to Longitude: 77° 34’ 13.51” E. The bacterial strain were isolated from the 10.0 gm sludge sample added in minimal salt media (MSM) 100 ml which contained (g/l) CaCl2 0.01; peptone 5.0; Na2HPO4 2.4; D-glucose 10.0; NH4NO3 0.1; K2HPO4 2.0; MgSO4 0.01, trace element (1 ml l−) and then adjusted pH 7.5±0.124,25. The flasks were incubated at 37 ± 1 °C in incubator shaker (Orbitek, Scigenic India) for 7 days at 120 rpm. Subsequently, an aliquot 1ml sample was serially diluted (10-1-10-8) with distilled water and 100μL diluted sample spread on MSM agar medium plates and incubated at 30°C. Finally, bacterial colonies were screened in the presence of guaiacol according to the method followed by DT Souza et al.26. The reddish-brown color halos showed a bacterial colony with laccase activity.
Molecular characterization
The identification of isolated bacterial strain was done from the Chromous Biotech Pvt. Ltd (Bangalore, India). The PCR amplicon of forward and reverse Prokaryotes 16S rRNA specific primer (PCR product size ~1.5kb) using 27F 5’–AGAGTTTGATCMTGGCTCAG–3’ forward primer and 1492R 5’–TACGGYTACCTTGTTACGACTT–3’ reverse primer by BDT v3.1 sequencing kit through ABI 3500xl Genetic Analyzer. The consensus analyzed sequence 1162 base pair of rDNA was produced by the use of aligner software in the forward and reverse sequence data. BLAST tool was used to search 16S rRNA sequence on the NCBI website in online mode (http:/www.ncbi.nlm.nih.gov/blast/). The chromatogram file verified both ends of the sequences, using the MEGABLAST algorithm to perform a Basic Local Alignment Search Tool (BLAST) with the NCBI Gene Bank database. According to Thompson et al.27 the multiple sequence alignment was carried out by using CLUSTALW and further, neighbor-joining method used to depict evolutionary history28.
Production of bacterial laccase
Preparation of agro-wastes substrate
Different types of agro-wastes (i.e. wheat bran, rice bran sawdust, banana peel, orange peel, pea peel, potato peel, and sugarcane bagasse) were collected around the local market Lucknow and dried at 60°C. After the removal of dust particles and moisture of these waste materials grinded in mixer grinder (Maharaja Mx-116A) and stored for further use in dry place29.
Screening of suitable agro-waste substrate
Suitable agro-waste substrates were screened for the maximum laccase production in submerged fermentation state. Two grams of each agro-waste substrates were taken in each flask with 100 mL of mineral basal salt solution (MBSS) moistened29,30 and then autoclave. After autoclave, the flasks were cool down then inoculated with 3.1× 106 CFU of Bacillus sp. AKRC01 and incubated for 24 h. The cultures were grown in the media and produced laccase through their enzymatic activity. Continuous sampling was performed to estimate bacterial growth and laccase activity. The cultured sample was extracted and centrifuged (10000rpm) at 4°C for 10 minutes after that the crude enzyme in the supernatant was used and pallet discorded.
Laccase assay
According to Arora et al.31, laccase activity was measured and the reaction mixture was incubated at 25°C for 2 hours. The reaction mixture contains 0.2 ml enzyme extract; 1 ml guaiacol (2 mM) and 3.8 ml 10 mM acetate buffer pH 5.0 were incubated at 25°C for 2 hours. The absorption was measured at 450 nm using UV-Vis spectrophotometer (Evolution 201) and laccase activity was evaluated in U/ml and defines as the quantity of enzyme that catalyzes one micromole per min oxidized substrate.
Experimental design using RSM-CCD
The physicochemical effects i.e. incubation time, temperature, and pH were optimized for the maximum enzyme production. The screened suitable agro-waste substrate for maximum enzyme production in submerged fermentation conditions (SmF) using the central composite design (CCD) process. However, incubation time, temperature and pH were the basic parameters for the optimization of the enzyme. Response surface designs such as CCDs are capable of fitting a complete quadratic model32-34. The effect of each factor for enzyme production was investigated at five separates concentrations: -α, -1, 0, + 1 and + α. Here, a separate set of 20 triplicates was also runs32,33,35. At a core coded value, all variables were considered to be zero. Variance analysis (ANOVA) was used to analyze the information collected from RSM in the production of laccase. The second-order polynomial Eq. 1 found a match using response surface methodology (RSM).
Y= b0 + b1A + b2B + b3C + b1 b1A2 + b2 b2B2 + b3 b3C2 + b1 b2AB + b1 b3AC + b2 b3BC
Where, Y factor, b0 interactive coefficients, b1, b2, b3 linear coefficients, b1,1, b 2,2, b 3,3 square coefficients, b 1,2, b 1,3, b 2,3 coefficients, and A, B, C, A2, B2, C2, AB, AC, BC autonomous coefficients. The statistical software Design-Expert software-11 (student version) was used for analyzing the information, optimizing the data and creating 2D and 3D interactive graphs of the response surface.
Effects of supplementary carbon and nitrogen sources for enzyme production
In the culture medium, the nature and amount of carbon and nitrogen sources are essential for bacterial growth which increased the production of extracellular laccase enzymes. The production medium was enriched with different concentrations (1.0% 1.5 % and 2.0%) of carbon source (glucose, sucrose, maltose, starch, and lactose) depend upon the carbon chain and structural difference. Nitrogen source used on the basis of percentage and structural liberty (peptone, urea, and yeast extract, ammonium sulfate and sodium nitrate) in 250mL Erlenmeyer flasks containing 50mL of MBSS medium with 2.0 g (v/w%) of rice bran. The flasks were sterilized, cooled at room temperature, and inoculated with 3.1× 106 CFU of Bacillus sp. AKRC01 and incubated at optimized environmental conditions with the agro-waste substrate. The contents of the flasks were centrifuged at 10000 rpm for 10min at 40C and the supernatant was used to assay for production of the enzyme at 450 nm.
Partial Purification and determination of molecular weight
In optimized conditions for laccase production was culture obtained and centrifuged at 8000 rpm at 4°C for 15 minutes. The supernatant was collected and using ammonium sulfate with continuous ice stirring and frozen in the ice of 2 hours for precipitation. The mixture was centrifuged at 10000 rpm for 10 min at 4°C and then mixed with 1.0 mL, 0.05mol L-1 acetate buffer having pH 5.0. All contents centrifuged again and dialyzed against 2L same acetate buffer via method followed by Aslam et al.36. The dialyzed extract was purified through a DEAE sephadex column using 0.1–1.0 M NaCl by ion exchange chromatography31. The same buffer eluted the enzyme and collected 2 ml fractions at a flow rate of 0.5 ml/min-1. According to Laemmli37, described SDS-PAGE and analyzed the purified fractions containing laccase and identified their molecular weight compared with the protein ladder. The molecular weight of laccase was determined using 10% polyacrylamide gel through SDS-PAGE. Coomassie Brilliant BlueR-250 (Sigma, St. Louis, MO, USA) stained the gel to visualize the protein band.
Statistical analysis
Every experiment was done in triplicate and all results have represented the average of observations. Variance analysis (ANOVA) was carried-out using Design-Expert software-11 (student version) on the experimental data obtained in the present research.
Isolation and screening of laccase producing bacterial strains
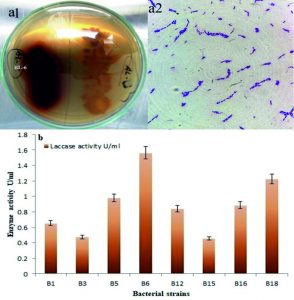
Morphologically different bacterial colonies were selected from the spread Petri plates of MSM agar media. The 26 bacterial strains were isolated and purified by the streak plate method in MSM agar media. All 26 bacterial strains were screened for the laccase activity on modified B and K agar medium containing 0.02% guaiacol that visualized as a reddish-brown color zone around the colonies. It is due to the guaiacol oxidation because guaiacol is the most convenient qualitative assay for laccase production by bacterial strains. Out of these, eight bacterial strains (i.e. B1, B3, B5, B6, B12, B15, B16, and B18) were showing the laccase activity Fig. 1 a1. The enzyme production studies were conducted in a submerged fermentation state and the highest enzyme producer was selected. Fig. 1b shows the maximum laccase production by isolates in 96 hours. Among the eight bacterial strains, the bacterial strain B6 was found to give the highest production laccase (1.564 U/ml) and was selected for further study. Further, selected bacterial strain B6 was examined under the microscope by gram’s staining (Fig. 1a 2). These bacteria were Gram’s positive, rod-shaped motile bacteria were observed under 100× magnification38.
Fig. 1. (a1) Laccase producing bacteria on B and K agar plate (a2) Gram’s staining (b) Screening of laccase producing bacterial strains.
Identification of laccase producing bacteria
The selected potential bacterial strain BL6 was identified through 16S rRNA sequencing. Sequence assessments among different species indicated the degree to which they are linked. This was performed using a neighbor-joining technique to construct the phylogenetic tree. The 16S rRNA sequence of bacterial strain Bacillus sp. AKRC01 was deposited in Gene Bank under the accession number MK478847. Moreover, 16S rDNA analysis of isolate AKRC01 (BL6) indicated 99% homology with Bacillus anthracis strain SS1 and 97% Bacillus sp. MH01. Although 16S rDNA sequencing does not provide authentic differentiation and identification Bacillus sp. AKRC01which is closely associated such as Bacillus cereus strain 67, Bacillus cereus strain QCG4. It can be used for initial associations (Fig. 2).
Agro waste substrate screening for laccase production in SmF
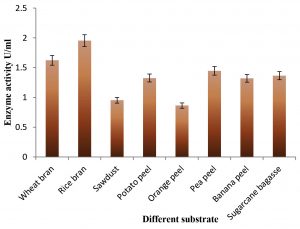
Exactly 2 g of each agro-waste substrate added in the different flask containing MBSS medium and inoculated 3.1× 106 CFU of Bacillus sp. AKRC01. Each flask having pH 7.0 was incubated for 120 hours and laccase production was measured at 24 hours intervals. Results of Fig. 3 showed that the maximum production of laccase (1.954 U/ml) was noted in the rice bran followed by wheat bran (1.625 U/ ml), sawdust (0.954 U/ml), potato peel (1.326 U/ml), orange peel (0.864 U/ml), pea peel (1.448 U/ml), banana peel (1.322 U/ml) and sugarcane bagasse (1.365 U/ml). Furthermore, studies were carried out using rice bran as an effective substrate for enzyme production in SmF. From this, it is evaluated that the agro wastes reduce the disposal problem and prevent probable environmental pollution.
Optimization by RSM
The three significant environmental parameters (incubation time, temperature and pH) that influenced the maximum production of laccase were used as central values to design experiments for central composite response surface design (Table 1). A predictive quadratic polynomial equation (2) using multiple regression analysis on the experimental data as shown in Fig.4a was designed to describe the actual and predictive correlation between laccase production and three significant parameters as follows:
Laccase production (U/ml) =-23.89210+0.098215 Time+0.453983 Tempereature+4.11600 pH+4.68750E-06 Time* Temperature -0.000078 Time* pH+0.004250 Temperearture* pH-0.000471 Time²-0.006991 Tempereature²-0.301163pH²
where, coded values are the A (incubation time), B (temperature), and C (pH). The variance analysis for the quadratic response surface model is presented in table 2. The Model F-value of 37.74 implies the model is significant. There is only a possibility (0.01%) that this large F-value might happen due to noise. P-values below 0.05 indicate significant terms for the model. In this case, A, A², B², C² were significant model terms. p values greater than (p>0.05) indicate the not significant model terms. In this case, A, A², B², C² were significant model terms. The determination coefficient R2 (0.8120) showed good compatibility between the values of experimental and predicted. The adjusted R2 value (0.9457) indicated that the enzyme activity variation (94.57%) was attributed to the independent variables and the model could not describe the total variation only (0.87%).
Fig. 4 (a). Three factors affecting laccase production of perturbation graph, (b) Relation between actual and predictive data, (c-h) 3D and 2D interaction plot of incubation time temperature and pH for laccase activity.
Table (1):
Central composite experiments design matrix with observed and predicted values of laccase production in rice bran by Bacillus sp. AKRC01.
| Factors | Rice bran | |||||
|---|---|---|---|---|---|---|
| Std | Run | A: Incubation time (hours) | B: Temperature (0C) | C: pH | Observed value | Predicted value |
| 8 | 1 | 48 | 25 | 5 | 0.354 | 0.432 |
| 20 | 2 | 144 | 25 | 5 | 1.202 | 1.342 |
| 9 | 3 | 48 | 45 | 5 | 0.452 | 0.398 |
| 7 | 4 | 144 | 45 | 5 | 0.645 | 0.698 |
| 19 | 5 | 48 | 25 | 9 | 0.524 | 0.643 |
| 13 | 6 | 144 | 25 | 9 | 0.678 | 0.78 |
| 15 | 7 | 48 | 45 | 9 | 0.298 | 0.342 |
| 10 | 8 | 144 | 45 | 9 | 1.125 | 1.24 |
| 6 | 9 | 24 | 35 | 7 | 0.214 | 0.236 |
| 4 | 10 | 168 | 35 | 7 | 1.823 | 1.976 |
| 3 | 11 | 96 | 20 | 7 | 2.023 | 2.02 |
| 18 | 12 | 96 | 50 | 7 | 1.752 | 1.65 |
| 5 | 13 | 96 | 35 | 4 | 0.456 | 0.454 |
| 17 | 14 | 96 | 35 | 10 | 0.987 | 0.923 |
| 1 | 15 | 96 | 35 | 8 | 3.832 | 3.986 |
| 14 | 16 | 96 | 35 | 7 | 3.452 | 3.654 |
| 12 | 17 | 96 | 35 | 7 | 3.457 | 3.53 |
| 11 | 18 | 96 | 35 | 7 | 3.565 | 3.64 |
| 2 | 19 | 96 | 35 | 7 | 3.645 | 3.78 |
| 16 | 20 | 96 | 35 | 7 | 3.512 | 3.43 |
Perturbation plot
The perturbation plot demonstrates how the response change as each factor passes from the selected reference point, with other variables at the reference value being kept constant (Fig. 4a). Laccase production was affected by the incubation period followed by temperature and pH (Fig. 4). Increasing the incubation period up to 96 hours resulted in elevated laccase production while increasing the temperature (35°C) and pH (8.0) up to the coded reference point (0.21) also increases the laccase productions. However, the increase in these parameters beyond the coded point may decrease the laccase production.
Table (2):
Analysis of variance (ANOVA) for laccase production in second-order polynomial model.
| Source | Sum of Squares | df | Mean Square | F-value | p-value | |
|---|---|---|---|---|---|---|
| Model | 34.08 | 9 | 3.79 | 37.74 | < 0.0001* | significant |
| A-Time | 1.57 | 1 | 1.57 | 15.68 | 0.0027 | |
| B-Temperature | 0.0332 | 1 | 0.0332 | 0.3312 | 0.5777 | |
| C-pH | 0.0854 | 1 | 0.0854 | 0.8506 | 0.3781 | |
| AB | 0.0000 | 1 | 0.0000 | 0.0004 | 0.9844 | |
| AC | 0.0005 | 1 | 0.0005 | 0.0045 | 0.9479 | |
| BC | 0.0578 | 1 | 0.0578 | 0.5760 | 0.4654 | |
| A² | 12.13 | 1 | 12.13 | 120.92 | < 0.0001* | |
| B² | 5.03 | 1 | 5.03 | 50.17 | < 0.0001* | |
| C² | 14.59 | 1 | 14.59 | 145.43 | < 0.0001* | |
| Residual | 1.00 | 10 | 0.1003 | |||
| Lack of Fit | 0.9774 | 6 | 0.1629 | 24.95 | 0.0039 | significant |
| Pure Error | 0.0261 | 4 | 0.0065 | |||
| Cor Total | 35.09 | 19 | ||||
| Std. Dev. | 0.3168 | |||||
| Mean | 1.70 | |||||
| C.V. % | 18.64 | |||||
| R² | 0.9714 | |||||
| Adjusted R² | 0.9457 | |||||
| Predicted R² | 0.8120 | |||||
| Adeq Precision | 15.9472 |
* significant
Diagnosis of the statistical model
The normal residual plot can analyze the statistical properties of the model and data points should be nearly linear. A non-linear pattern in the error (such as an S-shaped curve) denotes non-normality that can be adjusted by a conversion. The residual from the production of laccase indicates the normal distribution (Fig. 4b). No data abnormalities are indicated by their alignment on the drawn line.
Interaction between variables
When the third is kept at its optimum value, the interaction between two factors was shown in 3D and 2D graphs (Fig. 4c-h). The 3D and 2D graphs showed how the production of laccase influenced by the variability of two variables at a time, maintaining the other in optimum conditions. Laccase enzyme production increased with increase optimum incubation time up to (96 hours). Incubation time, temperature and pH were played a major role in enzyme production in SmF conditions (Fig. 4c-d). Moreover, initial water amounts adversely impacted the microbial activity. This may be because the porosity of the medium reduces as the liquid content increases leading to easy oxygen transfer37. According to Niladevi et al.33,34, different variables fermentation such as incubation time, temperature and pH were found to have a major role for laccase production by Streptomyces psammoticus.
Qualitative optimization of carbon and nitrogen sources using RSM Optimized Value
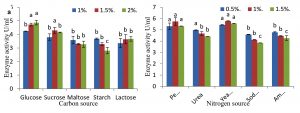
In present study physical factors, optimized value (incubation period 96-hour, temperature 35°C and pH 8.0) were used for further qualitative optimization of carbon sources (glucose, fructose, maltose, starch, and sucrose) and nitrogen source (peptone, urea, yeast extract, and ammonium sulfate, sodium nitrate). The use of different concentrations of carbon and nitrogen source for laccase production in SmF has also been well-documented39-41. In this optimization process, the most influencing additional nutrient source affected laccase production and combined with glucose 2.0% and peptone 1.0 %. Optimized nutritional conditions with 2.0% glucose (4.967 U/ml) and 1.0% peptone (6.236 U/ml) supplements gains maximum production of extracellular bacterial laccase (Fig. 5a and 5b). According to Muthukumarasamy et al.30, they reported that 3% of sucrose showed the maximum activity with rice bran. Similarly, nitrogen source and peptone showed higher activity with rice bran34. Annova was used to examine the experiment results and to evaluate how many differences each variable contributed.
Fig. 5. Effect of (a) carbon and, (b) nitrogen sources on laccase activity using rice bran as a substrate.
Validation of the model
The statistical tool of response surface model for all experimental design has been validated. To study laccase optimization, a random set of the experimental combination was used. The validation experiment clearly demonstrates that the experimental production value of 6.236 U/ml and predicted value 5.935 U/ml was in close agreement with the statistically. The results validated that the preceding model with different parameters such as incubation time (96 hours), temperature (35°C), pH (8.0) and additional carbon (2.0%) and nitrogen sources (1.0%) was the optimum conditions for maximum laccase production, confirming the model’s precision.
Purification of laccase and molecular weight determination
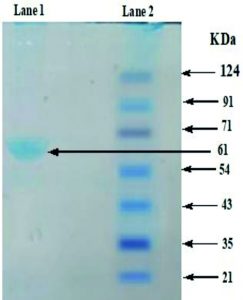
Extracellular laccase was purified from a culture of Bacillus sp. AKRC01 in optimized conditions for enzyme production. The laccase enzyme was purified from bacterial culture and determined molecular weight by denaturing 10% SDS-PAGE performed. The molecular weight of laccase produced by Bacillus sp. AKRC01 in the presence of rice bran was found to be 61 kDa which is inconsistent with the previous research (Fig. 6). Niladevi et al.33 discovered that Streptomyces psammoticus, unusual halotolerant-alkaline laccase having a molecular weight of about 43kDa and Bacillus subtilis WPI was also reported for a laccase production and determined the molecular weight of 55 kDa42. Earlier, a purified CotA type laccase protein from Bacillus pumilus, with an estimated 58 kDa molecular weight were also reported43.
The purpose of this study for maximum production of bacterial laccase enzyme in submerged fermentation state is a viable approach to the cost-related problems encountered in the production of enzymes. The optimized environmental factors (incubation period 96 h, temperature 35 °C and pH 8.0) and nutritional conditions (glucose 2.0% and peptone 1.0%) are the crucial parameters enhancing the maximum laccase production (6.236 U/ml) by Bacillus sp. AKRC01 in the presence of rice bran as an effective agro-waste substrate. The production of laccase by bacteria has been broadly studied due to their secretion of enzyme and their growth using agro-waste as a cheap substrate. Using agro-waste as a substrate not only reduces the disposal problem but also minimizes the risk of environmental pollutions.
ACKNOWLEDGMENTS
Authors would like to thank the Babasaheb Bhimrao Ambedkar University and Prof. (Dr.) Ram Chandra for providing the facilities to undertake this research work.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
Mr. AK and AK Singh has done experimental work and drafting of the manuscript, Mr. SA has supported for the data analysis of RSM while, RC has supervised the work and final correction of the manuscript.
FUNDING
The financial assistance from DBT, New Delhi, India Letter Number BT/PR18896/BCE/8/1372/2016 dated 28-03-2018 to Prof. Ram Chandra and University Grant Commission, New Delhi, to Adarsh Kumar Ph.D. scholar, is highly acknowledged and NFPwD (F No. 01-01/2019-Sch.) to Ajay Kumar Singh.
ETHICS STATEMENT
Not applicable.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
- Wong Y, Yu J. Laccase-catalyzed decoiorization of synthetic dyes. Water Res., 1999; 33: 3512-3520.
Crossref - Mendoza L, Jonstrup M, Hatti-Kaul R, Mattiasson B. Azo dye decolorization by a laccase/mediator system in a membrane reactor: enzyme and mediator reusability. Enzyme Microb. Technol., 2011; 49(5): 478-484.
Crossref - Mayer AM, Staples RC. Laccase: new functions for an old enzyme. Phytochemistry, 2002; 60: 551-565.
Crossref - Nelson D, Elisa E. Potential applications of oxidative enzymes and phenol-oxidase-like compounds in wastewater and soil treatment. Appl. Catal. B-Environ., 2000; 28: 83-99.
Crossref - Nelson D, Maria AR, Alessandro D, Liliana G. Applications of laccases and tyrosinases (phenoloxidases) immobilized on different supports. Enzyme Microb. Tech., 2002; 31: 907-931.
Crossref - Jhadav A, Vamsi KK, Khairnar Y, Boraste A, Gupta N, Trivedi S, Patil P, Gupta G, Gupta M, Mujapara AK, Joshi B, Mishra D. Optimization of production and partial purification of laccase by Phanerochaete chrysosporium using submerged fermentation. International Journal of Microbiology Research, 2009; 1: (2) 09-12.
- Narayanan PM, Murugan S. Production, purification and application of bacterial laccase: a review. Biotechnology, 2014; 13: 196–205.
Crossref - Baldrian P, Snajdr J. Production of ligninolytic enzymes by litter-decomposing fungi and their ability to decolorize synthetic dyes. Enzyme and Microbial Technology, 2006; 39(5): 1023–1029.
Crossref - Couto SR, Sanroman MA. Application of solid-state fermentation to ligninolytic enzyme production, Biochemical Engineering Journal, 2005; 22(3): 211–219.
Crossref - Couto SR, Herrera JLT. Industrial and biotechnological applications of laccases: a review, Biotechnology Advances, 2006; 5(24): 500–513.
Crossref - Revankar MS, Lele SS. Enhanced production of laccase using a new isolate of white rot fungus WR-1. Process Biochemistry, 2006; 41(3): 581–588.
Crossref - Chawachart N, Khanongnuch C, Watanabe T, Lumyong S. Rice bran as an efficient substrate for laccase production from thermotolerant basidiomycete Coriolus versicolor strain RC3. Fungal Diversity, 2004; 15(2): 23–32.
- Lorenzo M, Moldes D, Couto SR, Sanroman A. Improving laccase production by employing different lignocellulosic wastes in submerged cultures of Trametes versicolor. Bioresource Technology, 2002; 82(2): 109–113.
Crossref - Nyanhongo GS, Gomes J, Gubitz G, Zvauya R, Read JS, Steiner W. Production of laccase by a newly isolated strain of Trametes modesta. Bioresource Technology, 2002; 84(3): 259–263.
Crossref - Govarthanan M, Park SH, Kim J, Lee K, Cho M, Kamala-Kannan S, Oh B. Statistical optimization of alkaline protease production from brackish environment Bacillus sp. SKK11 by using horse gram husk. Preparative Biochemistry and Biotechnology, 2014; 44(2): 119-131.
Crossref - Govarthanan M, Selvam, Kandasamy, Munisamy, Govindaraju, Kamalakannan, Seralathan, Balakrishnan, Senthilkumar, Thangaswamy, Selvankumar. Process optimization of cellulase production from alkali-treated coffee pulp and pineapple waste using Acinetobacter sp TSK-MASC. RSC Advances, 2014; 4(25): 13045-13051.
Crossref - Selvam, Kandasamy, Thangaswamy, Selvankumar, Rajiniganth, Radhika, Srinivasan, Palanisamy, Sudhakar, Chinnappan, Balakrishnan, Senthilkumar, Muthusamy, Govarthanan. Enhanced production of amylase from Bacillus sp. using groundnut shell and cassava waste as a substrate under process optimization: Waste to wealth approach. Biocatalysis and Agricultural Biotechnology, 2016; 7; 250-256.
Crossref - Muthusamy, Govarthanan, Thangaswamy, Selvankumar, Selvam, Kandasamy, Sudhakar, Chinnappan, Aroulmoji, Vincent, Kamalakannan, Seralathan. Response surface methodology based optimization of keratinase production from alkali-treated feather waste and horn waste using Bacillus sp. MG-MASC-BT. Journal of Industrial and Engineering Chemistry, 2015; 27.
Crossref - Boza A, De la Cruz Y, Jordan G, Jauregui-Haza U, Aleman A, Caraballo I. Statistical optimization of a sustained release matrix tablet of lobenzarit disodium. Drug. Dev. Ind. Pharm., 2000; 26: 1303-1307.
Crossref - Singh SK, Dodge J, Durrani MJ, Khan MA. Optimization and characterization of controlled release pellets coated with experimental latex: I. Anionic drug. Int. J. Pharm., 1995; 125: 243-255.
Crossref - Sanchez-Lafuente C, Furlanetto S, Fernandez-Arevalo M. Didanosine extended-release matrix tablets: Optimization of formulation variables using statistical experimental design. Int. J. Pharm., 2002; 237: 107-118.
Crossref - Francis F, Sabu A, Nampoothiri KM, Ramachandran S, Ghosh S, Szakacs G. Use of response surface methodology for optimizing process parameters for the production of a- amylase by Aspergillus oryzae. Biochem. Eng. J., 2003; 107-115.
Crossref - Srinivasan, Palanisamy, Thangaswamy, Selvankumar, Kamalakannan, Seralathan, Mythili, Raja, Sengottaiyan, Arumugam & Muthusamy, Govarthanan & Balakrishnan, Senthilkumar & Selvam, Kandasamy. Production and purification of laccase by Bacillus sp. using millet husks and its pesticide degradation application. 3Biotech, 2019; 396.
Crossref - Chandra R, Raj A, Purohit HJ, Kapley A. Characterization and optimization of three potential aerobic bacterial strains for kraft lignin degradation from pulp paper waste. Chemosphere., 2007; 67: 839–846.
Crossref - Pfenning N, Lippert KD. Uber das vitamin B-12- bedrurbins phototropher schwefelbakterien. Archives of Microbiology, 1966; 55: 245–256.
Crossref - DT D’Souza, Tiwari R, Sah AK, Raghukumar C. Enhanced production of laccase by a marine fungus during treatment of colored effluents and synthetic dyes, Enzyme and Microbial Technology, 2006; 38: 504–511.
Crossref - Thompson JD, Higgins DG, Gibson TJ, Clustal W. Improving the Sensitivity of Progressive Multiple Sequence Alignment Through Sequence Weighting, Position-Specific Gap Penalties and Weight Matrix Choice. Nucleic. Acids. Res., 1994; 22: 4673–4680.
Crossref - Saitou N, Nei M. The Neighbor-Joining Method: A New Method for Reconstructing Phylogenetic Trees”. Mol. Biol. Evol., 1987; 4: 406–425.
- Poonam SN, Pandey A. Biotechnology for Agro-Industrial Residues Utilisation: Utilisation of Agro-Residues. Springer Science and Business Media, 2009.
- Muthukumarasamy NP, Jackson B, Raj AJ, Sevanan M. Production of Extracellular Laccase from Bacillus subtilis MTCC 2414 Using Agroresidues as a Potential Substrate. Biochemistry Research International, 2015; ID 765190, 9.
Crossref - Arora DS, Chander M, Gill PK. Involvement of lignin peroxidase, manganese peroxidase and laccase in degradation and selective ligninolysis of wheat straw. International Biodeterioration & Biodegradation, 2002; 50: 115-120.
Crossref - Fatma G, Satinder KB, Tyagi RD, Rojan P, John M, Valero JR. Parameter optimization for production of ligninolytic enzymes using agro-industrial wastes by response surface method. Biotechnology and Bioprocess Engineering, 2011; 16: 343-351.
Crossref - Niladevi KN, Prema P. Immobilization of laccase from Streptomyces psammoticus and its application inphenol removal using packed bed reactor. World Journal of Microbiology and Biotechnology, 2008; 24: 1215–1222.
Crossref - Niladevi KN, Sukumaran RK, Prema P. Utilization of rice straw for laccase production by Streptomyces psammoticus in solid-state fermentation. J. Ind. Microbiol Biotechnol., 2007; 34(10) 665-674.
Crossref - Bains J, Capalash N, Sharma P. Laccase from a nonmelanogenic, alkalotolerantc-proteobacterium JB isolated from industrial wastewater drained soil. Biotechnol Lett., 2003; 25(1):155–1159.
Crossref - Aslam MS, Aishy A, Samra ZQ, Gull I, Athar MA. Identification, purification and characterization of a novel extracellular laccase from Cladosporium Cladosporioides. Biotechnology & Biotechnological Equipment., 2012; 26(6): 3345-3350.
Crossref - Laemmli UK. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature, 2007; 227: 680–685.
Crossref - Barrow GI, Feltham RKA. Cowan and Steel’s manual for the identification of medical bacteria 3rd Edn. Cambridge University., 1993.
Crossref - Patrick F, Mtui G, Mshandete AM, Kivaisi A. Optimization of laccase and manganese peroxidase production in submerged culture of Pleurotus sajorcaju. Afr. J. Biotechnol., 2011; 10: 10166–10177.
- Niladevi KN, Jacob N, Prema P. Evidence for a halotolerant- alkaline laccase in Streptomyces psammoticus Purification and characterization. Process Biochem., 2008; 43: 654–660.
Crossref - Nandal P, Ravella SR, Kuhad RC. Laccase production by Coriolopsis caperata RCK2011: optimization under solid state fermentation by Taguchi DOE methodology. Sci. Rep., 2013; 3: 1–7.
Crossref - Sheikhi F, Ardakani MR, Enayatizamir N, Rodriguez-Couto S. The Determination of Assay for Laccase of Bacillus subtilis WPI with Two Classes of Chemical Compounds as Substrates. Indian J Microbiol., 2012; 52(4): 701–707.
Crossref - Reiss R, Ihssen J, Thony-Meyer L. Bacillus pumilus laccase: a heat stable enzyme with a wide substrate spectrum. BMC Biotechnol., 2011; 11: 1–11.
Crossref
© The Author(s) 2020. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.