ISSN: 0973-7510
E-ISSN: 2581-690X
Yeasts are prevalent on the surfaces and within the pulp of ripening fruits, where they utilize available sugars and nutrients to thrive. This study focuses on the endemic Lai fruit (Durio sp.) from Kalimantan, Indonesia, which contains pectin, a key substrate for pectinolytic yeasts. The activity of these yeasts, particularly in producing pectinase enzymes, is influenced by the carbon and nitrogen resources present in the fruit. We aimed to evaluate the effects of varying carbon sources (glucose, sucrose, and pectin) and nitrogen sources (peptone, urea, and ammonium chloride) on the pectinase activity of yeasts isolated from Lai fruit. In our screening, five out of twenty yeast isolates demonstrated pectinase production, with three isolates UNJCC Y-144, UNJCC Y-148, and UNJCC Y-149 selected for further analysis. The initial screening revealed pectinolytic index values of 0.55 ± 0.07 mm, 0.45 ± 0.02 mm, and 0.49 ± 0.06 mm for UNJCC Y-144, UNJCC Y-148, and UNJCC Y-149, respectively. Molecular identification confirmed that UNJCC Y-144 is Cyberlindnera fabianii (100% homology), while UNJCC Y-148 and UNJCC Y-149 are Candida tropicalis (100% and 99.83% homology, respectively). Notably, Cyberlindnera fabianii UNJCC Y-144 exhibited the enzyme activity (EA) in a medium lacking pectin, ranging from 48.68 ± 2.84 to 32.05 ± 2.46 U/mL. These findings underscore the potential of utilizing local yeasts for biotechnological applications in enzyme production, particularly in the food industry, where pectinase plays a critical role in processing fruit and improving juice extraction efficiency.
Carbon and Nitrogen Sources, Pectinase Production, Solid-state Fermentation, Cyberlindnera fabianii, Candida tropicalis, Lai Fruit
Pectin is a structural component of the middle lamella in plant walls as well as one of the main elements of cereals, vegetables, fruits, and fiber.1 Pectin plays a role in the mechanical strength and physical properties of the primary cell wall in maintaining the structural integrity of the fruit.2 Pectin consists of galacturonan and rhamnogalacturonan3 and can be degraded by an enzyme, namely the pectinase enzyme.
Pectinase has various roles in industry, including as a biological catalyst in industries such as extraction and clarification of fruit juices in food industries.4,5 It is also used for biotechnology and is needed for up to 25% of the demand in the food and beverage industry sector.6 Pectinase plays a role in the degradation of agricultural wastes,7 helps the fermentation process of tea and coffee,8 catalyzes the process of bleaching paper, the process of forming textile yarns, and modifying the pectin substance during the fruit ripening process.9 The pectinase enzyme can degrade the homogalacturonan and rhamnogalacturonan regions of pectin.10
Naturally, pectinase can be found in animals, plants, and microorganisms. Currently, microorganisms are the primary source of pectinase because of their easy cultivation, more straightforward enzyme harvesting process, and the final yield that can be improved through genetic engineering.11 Pectinase is produced naturally in microorganisms and has been widely isolated from fungi such as Aspergillus indicus, A. flavus, A. niveus.12 Besides fungi, yeasts also have the ability to produce pectinase enzymes, including Saccharomyces fragilis, Torulopsis kefir, Kluyveromyces marxianus and S. cerevisiae.12-14
One of the most common substrates for pectinase-producing yeasts is fruit. Fruit is a suitable substrate for the growth of various types of yeast because of the relatively high sugar content and other nutrients.15-17 Several studies have explored yeast diversity in fruits: The isolation of yeast species like Kloeckera apis, Saccharomyces cerevisiae, and Candida tropicalis from cocoa fruit fermentation is particularly noteworthy.18 These yeasts likely play crucial roles in breaking down the complex carbohydrates and other nutrients present in the fruit, contributing to the unique flavors and characteristics of the final fermented product. Even more intriguing is the discovery of pectinolytic yeasts, such as Candida parapsilosis, C. glabrata, and Rhodotorula mucilaginosa, in orange molasses.19 The ability of these yeasts to produce enzymes that degrade pectin, a major structural component of fruit cell walls, suggests they may have a competitive advantage in fruit-based environments. Their pectin-degrading capabilities could allow them to access and utilize a wider range of nutrients, potentially outcompeting other microorganisms.
Lai fruit is an endemic fruit typical of Kalimantan, Indonesia, which is similar to durian fruit in general but has a different texture and taste.20,21 Lai fruit contains 2.92% protein, 34.69% carbohydrates, and 1.70% fat. The pectin content of tropical and subtropical fruits is a fascinating aspect of their composition and texture. According to the research, these fruits typically contain pectin levels ranging from 13-30%.22 This significant presence of pectin plays a crucial role in influencing the softness and overall mouthfeel of the fruits. The connection between pectin content and fruit softness lies in the activity of pectinase enzymes. As highlighted, pectinase activity is a key factor in the breakdown of pectin, the structural component of plant cell walls.23 When pectinase enzymes act on pectin, they facilitate the softening of the fruit’s texture, allowing it to become more supple and yielding to the touch.
The ability of yeast to produce pectinase enzymes is affected by various factors, including optimum nutrition availability such as carbon to increase biosynthetic energy24 and nitrogen as it is needed for the cell growth.25 Reported that Kluyveromyces marxianus uses galactose to produce pectinase enzyme with the activity value of 2.0 U/mL.26,27 This type of yeast with an additional carbon source of 1% citrus pectin and nitrogen source of peptone has a pectinase activity value of 4.2 U/mL.
The purpose of this research is to optimize pectinase enzyme synthesis by investigating the effects of adding various carbon and nitrogen sources to the growth medium of Cyberlindnera fabianii UNJCC Y-144 and Candida tropicalis UNJCC Y-148-149, both of which were isolated from Lai fruit. This approach not only advances our understanding of microbial enzyme production but also provides valuable insights into the optimal growth conditions for these specific yeast strains, potentially paving the way for more sustainable and efficient biotechnological.
Screening of yeast strains
Twenty representative yeast isolates were collected from Universitas Negeri Jakarta Culture Collection (UNJCC). The yeasts were isolated from Lai fruit, which has a high sugar content and soft texture. Saccharomyces cerevisiae UNJCC Y-87 was also used as a compared strain in this study. On a rotary shaker set at 180 rpm for 24 hours at 28 °C, yeast is grown on Peptone Dextrose Yeast Extract (10 g/L yeast extract, 20 g/L peptone, and 20 g/L dextrose) and Nutrient Yeast Dextrose Broth (8 g/L nutrient broth, 5 g/L yeast extract, and 10 g/L glucose). The method began with centrifuging the yeast cell suspension at 4000 x g for 10 minutes at 4 °C to separate cells from the growth medium. Following centrifugation, the medium was removed, and the cells were washed with distilled water to eliminate residual nutrients. The yeast cells were then resuspended to a final concentration of 5 x 108 cells/mL, ensuring a standardized cell density for subsequent analysis. Colonies exhibiting typical yeast morphology were stored in the Universitas Negeri Jakarta Culture Collection (UNJCC). They were preserved at -20 °C using a 10% glycerol solution and subjected to lyophilization to ensure long-term stability and viability for future research use.
Screening of pectinase production
The screening of pectinase production was conducted using the method outlined by Sukmawati et al.14,28 with modification. Pectinase screening agar medium (PSAM) was employed to assess the ability of yeasts, incubated for 48 hours at a cell density of 107 CFU/mL, to produce the enzyme pectinase. The PSAM comprised 10 g/L of pectin, 1.4 g/L of (NH4)2 SO4, 6 g/L of K2HPO4, 2 g/L of KH2PO4, 0.1 g/L of MgSO4, and 20 g/L of agar, all prepared in a total volume of 1000 mL. To check for pectinolytic activity, the yeast isolate was incubated on pectin agar medium for 72 hours at 30 °C (Figure 1). The pectinolytic index value is calculated by measuring the effectiveness of yeast isolates in hydrolyzing pectin, which is indicated by a clear zone surrounding the colony on a pectinase screening agar medium. The pectinolytic index (PI) is determined using the following formula:
Pectinolytic Index (PI) = Dc / Dc + Dz
Where
Dc = Diameter of the colony (measured in millimeters).
Dz = Diameter of the clear zone surrounding the colony (measured in millimeters).
To calculate the pectinolytic index, the diameters of both the colony and the clear zone are measured using an analytical caliper. The values are then substituted into the formula to obtain the PI, which indicates the relative pectinase activity of each yeast isolate. A higher PI value signifies greater pectinolytic activity.
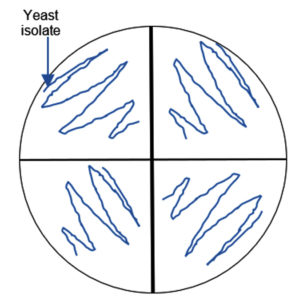
Figure 1. Semi-quantitative screening of pectin degrading yeasts. 20 mL of the 48-hour-incubated yeast cell suspension was inoculated into the wells in each quadrant
Macroscopic and microscopic identification of yeast
The identification of yeast species was conducted through both macroscopic and microscopic examinations of the colony morphology. Macroscopic observations included analyzing characteristics such as texture, color, surface appearance, profile, and colony edge. These features were assessed to determine distinctive traits that could help differentiate between yeast strains. Microscopic examination was performed using a phase-contrast microscope (Olympus) at 400x magnification, which allowed detailed observation of cell structure and budding patterns. This step was crucial for identifying yeast cell shapes and budding types, providing a finer level of detail to support accurate species identification.
Molecular identification of yeast isolates using D1/D2 region of 26S rDNA
DNA isolation of yeasts was carried out using the boiling method based on another research with modification.29 A full loop of yeast was inoculated onto YMA (Yeast Mold Agar) slanted medium and incubated for 48 hours at 28 °C, ensuring optimal growth conditions. After the incubation period, three loops of the cultured yeast were carefully transferred to a 1.5 mL Eppendorf tube containing 250 µl of ultrapure distilled water. The tube was gently mixed to create a homogenous suspension of the yeast cells for further analysis. After vortexing the yeast suspension for one minute to ensure thorough homogenization, it was transferred to a glass beaker containing water. The beaker was then incubated at 100 °C for 25 minutes to inactivate enzymes and extract genomic DNA. Following this step, the tube was removed, and the sample was subjected to centrifugation at 13,000 rpm for 15 minutes to separate the supernatant from any remaining cellular debris.
The supernatant obtained from the centrifugation was used for the PCR (Polymerase Chain Reaction) analysis. A PCR cocktail was prepared, and the D1/D2 region of the LSU (Large Subunit) rRNA gene was amplified using the genomic DNA extracted from the yeast isolates. Forward primer NL1 (5′-GCATATCAATAAGCGGAGAAAG-3′) and reverse primer NL4 (5′-GGTCCGTGTTTCAAGACGG-3′) were used for molecular identification.30 After extracting DNA according to the previous instructions, 3 µL of DNA template was added with nuclease-free water (8.5 µL), Go Taq Green Mastermix (12.5 µL), NL1 forward primer (0.5 µL), NL4 reverse primer (0.5 µL) for PCR reaction. The following conditions were used for the 35 cycles of the PCR: pre-denaturation (95 °C for 2 min), post-denaturation (95 °C for 30 sec), annealing (58.0 °C for 30 sec), elongation (72 °C for 1 min), final elongation (72 °C for 10 min), and extension (4 °C) for final condition. The PCR results were sent to First Base DNA Sequencing Services for sequence extraction. Prior to this, the PCR products were visualized using electrophoresis on a 1% agarose gel in 1 x TAE (Tris-Acetate-EDTA) buffer, following the protocol outlined by Sambrook and Russell. After editing the sequencing data with ChromasPro version 2.6.2, the closest comparable species to the yeast isolates were identified using the Basic Local Alignment Search Tool (BLAST) available at http://www.ncbi.nlm.nih.gov. A phylogenetic tree was constructed using the neighbor-joining method with 1000 bootstrap replicates, employing the MEGA 7 software.31
Optimization of carbon and nitrogen addition for lab-scale enzyme production in solid-state fermentation
The inoculum used for the enzyme activity test was prepared according to the protocol established.14 Yeast isolates were transferred to YMA (Yeast Mold Agar) slant medium using sterile skewers, with a total of 15 strokes. The cultures were then incubated at 30 °C for 48 hours to promote growth. After incubation, the yeast isolate was homogenized, and 5 mL of sterile distilled water was added to create a yeast suspension. The optical density (OD) of the yeast suspension was then adjusted to 1.0 at a wavelength of 600 nm using a spectrophotometer. Optimization is based on carbon and nitrogen sources prepared with different fermentation media, based on variations in carbon and nitrogen sources. The carbon sources used were glucose, sucrose, and pectin with a carbon concentration of 1% (0.25 g in 25 mL medium). The nitrogen sources used were peptone, urea, and ammonium chloride with a concentration of 0.1% (0.025 g in 25 mL medium). The composition of the fermentation medium contains 1% carbon source, 0.1% nitrogen source, 0.6% K2HPO4, 0.20% KH2PO4 and 0.01% MgSO47H2O.
A 100 mL Erlenmeyer flask was used to create the fermentation medium, which had a medium volume of 25 mL and a pH of 5. Next, 10% (2.5 mL) of the yeast suspension was added to the fermentation medium along with different nitrogen and carbon sources. After that, the medium was incubated for 48 hours at 30 °C on a rotary shaker set to 150 rpm. After that, the crude enzyme extract, or supernatant, was centrifuged for 10 minutes at 4 °C at 3000 rpm. Enzyme activity was also assessed in the crude enzyme extract. The measured parameters were units/mg of a certain enzyme activity.
Calculation of pectinase enzyme activity using the DNS method
Using the dinitrosalicylic (DNS) approach, the quantity of reducing sugar generated during the enzymatic hydrolysis of pectin was measured in order to determine the activity of the pectinase enzyme.32 500 µL of crude enzyme extract from each sample was combined with 0.5 mL of 0.1 M acetate buffer pH 5 and 500 µL of 1% pectin solution. After that, this solution was incubated for 15 minutes at 40 °C. 1000 µl of dinitrosalicylic acid reagent (DNS) was applied to halt the reaction after incubation. It was then followed by incubation at 100 °C for 10 min and was cooled.
After the sample had cooled, its absorbance was measured at 540 nm using a spectrophotometer. The concentration of reduced sugars released was determined using a glucose standard curve. Enzyme activity (EA) was quantified as micromoles of glucose released per minute per milliliter of enzyme (µmol/min/mL), following the method described.33 The pectinase enzyme activity was calculated using the following equation, as outlined by Anggraini et al.7
EA (U/mL) = {[(μg/mL of glu) / mol. wt of glu] / Time (min) x vol. of enzyme (mL) } x Dil. Factor of enzyme
Isolation and screening of pectin-degrading yeasts from Lai fruit (D. kutejensis)
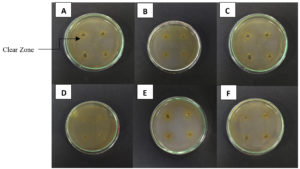
About 20 morphologically distinct pectinolytic colonies were isolated from Lai durian on YMA medium with white color, smooth colony surface, 70% mucoid texture, rough colony surface, and 30% butyrous texture (Figure 2). Research conducted reported that yeast isolated from Lai fruit were predominantly non-pigmented white colonies.29 Out of the 20, five isolates showed a significant clear zone around the colony by semi-quantitative screening, namely UNJCC Y-144, UNJCC Y-148, UNJCC Y-149, UNJCC Y-150, and UNJCC Y-151 (Figure 3). The growth of isolates is accompanied by an orange clearance zone, which indicates pectin hydrolysis. This clearance zone results from the production of galacturonic acid, a byproduct of pectin hydrolysis, which contributes to the formation of a brownish coloration in the surrounding medium. The five isolates’ cell-free extract revealed a distinct zone surrounding the inoculation wells. Thus, the enzyme can be considered extracellular.
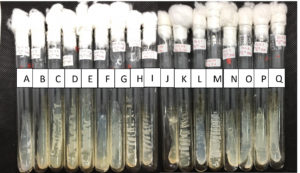
Figure 2. Yeasts collection from Universitas Negeri Jakarta Culture Collection (UNJCC). Yeasts were isolated from Lai fruit and incubated on YMA, at 28 °C for 48 h. From left to right: (A) Du 4.3; (B) Du 4.4; (C) Du 4.10; (D) UNJCC Y-144; (E) UNJCC Y-148; (F) Du 4.20; (G) Du 4.24; (H) UNJCC Y-149; (I) UNJCC Y-150; (J) Du 5.3; (K) Du 5.5; (L) Du 5.6; (M) UNJCC Y-151; (N) Du 6.2; (O) Du 6.3; (P) Du 6.4; (Q) Du 6.7
Figure 3. A clear zone around the colony marked screening of yeast isolates producing pectinase enzymes. (A) Saccharomyces cerevisiae UNJCC Y-87 (Control); (B) UNJCC Y-144; (C) UNJCC Y-148; (D) UNJCC Y-149; (E) UNJCC Y-150; (F) UNJCC Y-151 on Pectinase Screening Agar Medium, incubation at 30 °C for 72 h
The pectinolytic index (PI) is calculated as the ratio of the diameter of the clear zone to the diameter of the colony. The pectinolytic index values for the yeast isolates obtained from Lai fruit were analyzed using descriptive statistical methods. The results showed that isolate UNJCC Y-144 has the highest pectinolytic index value of 0.55 ± 0.07 mm. The pectinolytic index value of isolate UNJCC Y-144 was not significantly different from the isolate UNJCC Y-149 with a value of 0.49 ± 0.06 mm. This indicated that isolate UNJCC Y-144 and UNJCC Y-149 have the same pectinolytic index value (Table 1). The yeast isolate UNJCC Y-151 showed the lowest average pectinolytic index, measuring 0.25 ± 0.02 mm. In comparison, another research reported significantly higher pectinolytic index values, with Debaryomyces hansenii and Candida zeylanoides demonstrating indices of 4.3 mm and 5.3 mm, respectively.27 The three groups of yeast isolates, namely UNJCC Y-144, UNJCC Y-148, and UNJCC Y-149, with high pectinolytic index values, were identified and used in testing the optimization of pectinase enzyme activity based on carbon sources and nitrogen sources.
Table (1):
The pectinolytic index value of 5 yeast isolates on pectinase screening agar medium was incubated for 72 h at 30 °C. The pectinolytic index value defines the ratio between the diameter of the clear zone and the diameter of the colony
Isolate Code |
The pectinolytic index value (Mean ± SE) mm |
|---|---|
UNJCC Y-144 |
0.55 ± 0.07 |
UNJCC Y-148 |
0.45 ± 0.02 |
UNJCC Y-149 |
0.49 ± 0.06 |
UNJCC Y-150 |
0.32 ± 0.03 |
UNJCC Y-151 |
0.25 ± 0.02 |
Remarks: Numbers followed by the same letter are not significantly different at = 0.05 Duncan Multiple Range Test (DMRT)
Molecular identification and morphological characteristics of isolate UNJCC Y-144, UNJCC Y-148, and UNJCC Y-149
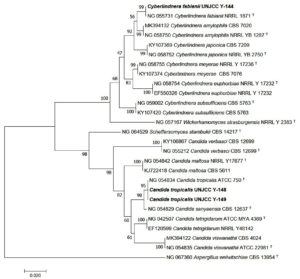
Molecular identification was conducted on three isolates, UNJCC Y-144, UNJCC Y-148, and UNJCC Y-149. The PCR process yielded nucleotide sequences approximately 600 base pairs in length. With a sequence homology of 100%, isolate UNJCC Y-144 is shown to have the highest homology to Cyberlindnera fabianii NRLL 1871. With a 100% sequence homology, isolate UNJCC Y-148 is most similar to Candida tropicalis ATCC 750 (Figure 4). With a sequence homology of 99.83%, isolate UNJCC Y-149 is most similar to Candida tropicalis ATCC 750 (Table 2).
Table (2):
BLAST results of UNJCC Y-144, UNJCC Y-148, and UNJCC Y-149 based on D1/D2 rDNA sequence analysis
Isolate Codes |
Identity |
Max Score |
Query Cover |
E-value |
Accession |
Identity |
Gaps (%) |
|---|---|---|---|---|---|---|---|
UNJCC Y-144 |
Cyberlindnera fabianii NRLL 1871 |
1072 |
95% |
0.0 |
NG_055731.1 |
100.00% |
0/580 (0%) |
UNJCC Y-148 |
Candida tropicalis ATCC 750 |
1101 |
96% |
0.0 |
NG_054834.1 |
100.00% |
0/596 (0%) |
UNJCC Y-149 |
Candida tropicalis ATCC 750 |
1094 |
93% |
0.0 |
NG_054834.1 |
99.83% |
0/582 (0%) |
Figure 4. Phylogenetic tree of isolates UNJCC Y-144, UNJCC Y-148, and UNJCC Y-149 based on the D1/D2 analysis with Neighbor-joining method (1000x Boostrap). The bootstrap value of above 95 indicates the tree is robust enough to use as a reference. Aspergillus welwitschiae CBS 13954 is used as an outgroup
Morphological characteristics of pectinolytic yeasts
The pectinolytic yeast Cy. fabianii UNJCC Y-144, C. tropicalis UNJCC Y-148, and C. tropicalis UNJCC Y-149 were characterized based on macroscopic and microscopic observation. The results showed that all three isolates have similar characteristics to a white colony with mucoid texture, smooth surface, flat edges, and mountainous colony profile (Table 3). The microscopic characteristic of Cy. fabianii UNJCC Y-144 has a spherical cell shape and monopolar budding with no hyphae. Meanwhile, C. tropicalis UNJCC Y-148 and C. tropicalis UNJCC Y-149 were round and oval cell shapes and monopolar buds with no hyphae. Suggested that the genus Candida has morphological characteristics of white to cream colonies, mucoid or dry texture, round to oval cells, and monopolar budding.34 This result is also confirmed by the statement that C. tropicalis has a smooth, white colony surface, the colony is round, and reproduces in monopolar budding.35
Table (3):
Macroscopic and microscopic observations of yeasts incubated on YMA medium for 48 h at 28 °C
Isolates |
UNJCC Code |
Color |
Texture |
Surface |
Edge |
Profile |
Cell Shape |
Budding |
Hyphae |
|---|---|---|---|---|---|---|---|---|---|
Cy. fabianii |
Y-144 |
White |
Mucoid |
smooth |
flat |
mountainous |
Round |
Monopolar |
– |
C. tropicalis |
Y-148 |
White |
Mucoid |
smooth |
flat |
mountainous |
Round and oval |
Monopolar |
– |
C. tropicalis |
Y-149 |
White |
Mucoid |
smooth |
flat |
mountainous |
Round and oval |
Monopolar |
– |
Calculation of relative enzyme activity based on carbon and nitrogen sources
The three-way ANOVA analysis revealed a significant effect of pectinase enzyme activity on the factors of isolate type, carbon source type, and nitrogen source, as indicated by a p-value of 0.00, which is less than 0.05. Additionally, a significant interaction was observed among these three factors, also with a p-value of 0.00 < 0.05. This result indicates that the value of the pectinase enzyme activity is influenced by the interaction between yeast isolate, the type of carbon source, and the type of nitrogen source, so a further DMRT test was carried out. Testing using the Duncan Multiple Range Test (DMRT) was done to see how treating isolates, carbon sources, and nitrogen sources affected the results. The results of the DMRT analysis revealed that the pectinase enzyme activity of Cyberlindnera fabianii UNJCC Y-144 in a medium containing urea and sucrose as carbon sources showed a significant difference at the five percent confidence level. This activity was notably distinct from other combinations of yeast isolates, carbon sources, and nitrogen sources, indicating that Cy. fabianii UNJCC Y-144’s enzyme production is especially responsive to this specific medium composition. Based on the results obtained, isolate Cy. fabianii UNJCC Y-144 has the maximum relative enzyme activity in medium added with sucrose and urea with the value of 48.68 ± 2.84 U/mL (Figure 5). The lowest relative enzyme activity is on a medium containing pectin and ammonium chloride with an activity value of 1.62 ± 0.25 U/mL. Isolate Cy. fabianii UNJCC Y-144 exhibited high relative enzyme activity in the medium not containing pectin as substrate, while it showed lower activity in the medium containing pectin as substrate molecules (Table 4).
Table (4):
Relative enzyme activity value of isolate Cy. fabianii UNJCC Y-144 using quantitative screening method
Carbon and Nitrogen Source |
OD at 540 nm |
mg/mL of glucose |
Enzyme Activity (U/mL) |
|---|---|---|---|
Peptone + glucose |
2.45 |
5.37 |
38.82 ± 0.89fg |
Peptone + sucrose |
2.66 |
5.84 |
42.27 ± 0.65gh |
Peptone + pectin |
0.36 |
0.75 |
4.57 ± 0.20a |
Urea + glucose |
2.73 |
6.00 |
43.48 ± 0.72h |
Urea + sucrose |
3.05 |
6.71 |
48.68 ± 2.84i |
Urea + pectin |
0.71 |
1.52 |
2.50 ± 0.25a |
Ammonium chloride + glucose |
2.75 |
6.05 |
43.78 ± 0.61h |
Ammonium chloride + sucrose |
2.04 |
4.46 |
32.05 ± 2.46de |
Ammonium chloride + pectin |
0.18 |
0.35 |
1.62 ± 0.25a |
Remarks: Values with the same letter are not significantly different at α = 0.05 (DMRT). ‘a’ indicates the lowest enzyme activity, while ‘de’, ‘fg’, ‘gh’, ‘h’, and ‘i’ show progressively higher levels.
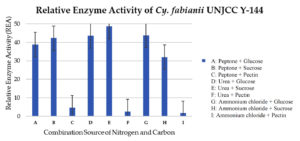 Figure 5. Relative enzyme activity of Cy. fabianii UNJCC Y-144. The standard error bars in the graph represent mean errors from duplicate samples tested
Figure 5. Relative enzyme activity of Cy. fabianii UNJCC Y-144. The standard error bars in the graph represent mean errors from duplicate samples tested
The maximum relative enzyme activity of C. tropicalis UNJCC Y-148 was obtained in the medium with glucose as carbon source and peptone as nitrogen source with an activity value of 10.63 ± 0.97 U/mL (Figure 6). It reached the lowest activity (0.12 ± 0.00 U/mL) when the isolate was cultivated on a medium containing ammonium chloride and sucrose. C. tropicalis UNJCC Y-148 exhibited significantly lower enzyme activity in all mediums compared to Cy. fabianii UNJCC Y-144 (Table 5).
Table (5):
Relative enzyme activity value of isolate C. tropicalis UNJCC Y-148 using quantitative screening method
Carbon and Nitrogen Source |
OD at 540 nm |
mg/mL of glucose |
Enzyme Activity (U/mL) |
|---|---|---|---|
Peptone + glucose |
0.29 |
0.59 |
10.63 ± 0.97b |
Peptone + sucrose |
0.20 |
0.40 |
0.27 ± 0.04a |
Peptone + pectin |
0.17 |
0.33 |
2.81 ± 0.031a |
Urea + glucose |
0.12 |
0.21 |
0.88 ± 0.14a |
Urea + sucrose |
0.20 |
0.38 |
0.27 ± 0.02a |
Urea + pectin |
0.17 |
0.32 |
2.63 ± 0.08a |
Ammonium chloride + glucose |
0.11 |
0.18 |
0.82 ± 0.03a |
Ammonium chloride + sucrose |
0.13 |
0.24 |
0.12 ± 0.00a |
Ammonium chloride + pectin |
0.19 |
0.36 |
1.10 ± 0.06a |
Remarks: Values with the same letter are not significantly different at α = 0.05 (DMRT). ‘a’ indicates the lowest enzyme activity, while ‘b’ indicates a significantly higher activity group.
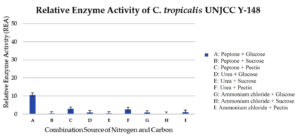 Figure 6. Relative enzyme activity of C. tropicalis UNJCC Y-148. The standard error bars in the graph represent mean errors from duplicate samples tested
Figure 6. Relative enzyme activity of C. tropicalis UNJCC Y-148. The standard error bars in the graph represent mean errors from duplicate samples tested
Isolate C. tropicalis UNJCC Y-149 exhibited fewer enzyme activity than the other isolated. The relative enzyme activity values in each combination medium range from 4.41 ± 0.64 to 0.13 ± 0.06 U/mL (Table 6). The maximum value obtained was in a medium containing glucose and peptone (Figure 7).
Table (6):
Relative enzyme activity value of isolate C. tropicalis UNJCC Y-149 using quantitative screening method
Carbon and Nitrogen Source |
OD at 540 nm |
mg/mL of glucose |
Enzyme Activity |
|---|---|---|---|
Peptone + glucose |
0.27 |
0.55 |
4.41 ± 0.64a |
Peptone + sucrose |
0.18 |
0.34 |
0.08 ± 0.03a |
Peptone + pectin |
0.24 |
0.49 |
3.90 ± 0.32a |
Urea + glucose |
0.11 |
0.20 |
0.66 ± 0.07a |
Urea + sucrose |
0.15 |
0.28 |
0.25 ± 0.01a |
Urea + pectin |
0.20 |
0.40 |
2.52 ± 0.05a |
Ammonium chloride + glucose |
0.11 |
0.19 |
0.67 ± 0.10a |
Ammonium chloride + sucrose |
0.13 |
0.24 |
0.13 ± 0.06a |
Ammonium chloride + pectin |
0.13 |
0.23 |
1.47 ± 0.25a |
Remarks: Numbers followed by the same letter are not significantly different at α = 0.05 based on DMRT. Values marked with ‘a’ indicate the lowest enzyme activity group.
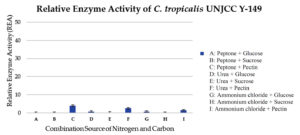 Figure 7. Relative enzyme activity of C. tropicalis UNJCC Y-149. The standard error bars in the graph represent mean errors from duplicate samples tested
Figure 7. Relative enzyme activity of C. tropicalis UNJCC Y-149. The standard error bars in the graph represent mean errors from duplicate samples tested
Isolate S. cerevisiae UNJCC Y-87 was used as a comparison isolate. In this test, the maximum value of relative enzyme activity was obtained in the medium with a sucrose carbon source and a urea nitrogen source with an activity value of 35.03 ± 5.62 U/mL (Figure 8). The lowest activity value was obtained in a medium with carbon sucrose and nitrogen source ammonium chloride, with a pectinase enzyme activity value of 0.57 ± 0.14 U/mL (Table 7). Isolate S. cerevisiae UNJCC Y-87 exhibited low enzyme activity in the medium containing pectin as substrate molecules (Figure 8).
Table (7):
Relative enzyme activity value of isolate S. cerevisiae UNJCC Y-87 using quantitative screening method
Carbon and Nitrogen Source |
OD at 540 nm |
mg/mL of glucose |
Enzyme Activity |
|---|---|---|---|
Peptone + glucose |
1.94 |
4.25 |
30.50 ± 4.25cd |
Peptone + sucrose |
1.73 |
3.77 |
26.95 ± 2.44c |
Peptone + pectin |
0.30 |
0.62 |
3.60 ± 0.30a |
Urea + glucose |
0.40 |
0.82 |
5.10 ± 0.44a |
Urea + sucrose |
2.22 |
4.87 |
35.03 ± 5.62ef |
Urea + pectin |
0.23 |
0.47 |
2.45 ± 0.15a |
Ammonium chloride + glucose |
0.15 |
0.28 |
1.05 ± 0.02a |
Ammonium chloride + sucrose |
0.12 |
0.21 |
0.57 ± 0.14a |
Ammonium chloride + pectin |
0.17 |
0.32 |
1.40 ± 0.16a |
Remarks: Values with the same letter are not significantly different at α = 0.05 (DMRT). ‘a’ indicates the lowest enzyme activity, ‘c’ and ‘cd’ indicate intermediate levels, and ‘ef’ indicates the highest enzyme activity.
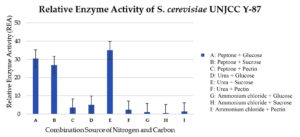 Figure 8. Relative enzyme activity of S. cerevisiae UNJCC Y-87. The standard error bars in the graph represent mean errors from duplicate samples tested
Figure 8. Relative enzyme activity of S. cerevisiae UNJCC Y-87. The standard error bars in the graph represent mean errors from duplicate samples tested
Cy. fabianii UNJCC Y-144 had the greatest enzyme activity value of 48.68 ± 2.84 U/mL in a medium containing a sucrose carbon source and urea nitrogen source, according to the optimisation of enzyme activity data. El-Enshasy et al. state that the pectinase enzyme, which had an enzyme activity value of 450 U/mL, can become more active throughout the fermentation process when a carbon source of sucrose is used.36 Yeast converts the disaccharide sucrose into glucose and fructose.
Yeasts are found in a wide range of substrates, including pectin-rich environments, as pectinases are commonly present in many higher plant species.19 The role of pectinase is to modify the pectin during fruit ripening.37 Pectinase can also be produced by microorganisms such as fungi and bacteria, which is considered more efficient than production from other sources.38 This can be seen from the cost-effectiveness, consistency, minimal time and space required for production, and ease of modification and optimization in the production process.10 The screening results showed that from 20 yeast isolates, 25% pectinolytic yeasts from Lai fruit were obtained, namely isolates UNJCC Y-144, UNJCC Y-148, UNJCC Y-149, UNJCC Y-150, and UNJCC Y-151 marked with a clear zone formed around the colony. The isolates that did not produce a clear zone around the colony indicated that the isolates could not produce pectinase enzymes. The clear zone formed around the colony indicated that the yeast isolates used pectin as a carbon source for their growth.
According to the greater the pectinolytic index, the greater the value of the enzyme activity produced.39 According fruit is a suitable substrate for various yeast species, including pectinolytic yeasts.17 Successfully isolated pectinolytic yeasts from fermented coffee cherries, including Pichia kudriavzevii and S. cerevisiae.40 The research conducted obtained a pectinolytic yeast Wickerhamomyces anomalus from citrus fruit.41 Obtained the pectinolytic yeast Kluyveromyces wickerhamii from the Brazilian tropical fruit, mangaba (Hancornia speciosa).42
In this study, the formation of a clear zone around colonies was observed after 72 hours of incubation on a pectinase screening agar medium. An iodine solution was then applied to highlight areas of pectinolytic activity, allowing for the detection of yeast strains with pectinase-producing capabilities.43 Iodine is firmly bound to a-1,4-glycosidic in the polysaccharides in selective pectin media.44 Yeast-produced metabolites clear zone formed is an area containing pectin which has been degraded by the pectinase enzyme produced by yeast. Suggested that the brownish color formed in the clear zone is the degradation of pectin that occurs by the pectinase enzyme produced by microbes.45
The calculation of the value of the pectinolytic index was carried out by the agar diffusion method. The agar diffusion method is easy and straightforward for measurement and analysis.46 In the agar diffusion method, 1% pectin concentration was used as the main carbon source in the pectinolytic screening test. The use of 1% pectin is the optimum concentration in this test. This is because if the pectin concentration is more than 1%, it can cause the medium to be too dense and inhibit the growth of yeast. High concentrations can increase the viscosity of the medium and inhibit the growth of microorganisms.47 Research conducted used 1% pectin concentration and obtained the optimum pectinolytic index value in yeasts Kluyveromyces wickherhamii and Pichia anomala.42
The results of semi-quantitative screening showed that the isolate UNJCC Y-144 has the highest average pectinolytic index value of 0.55 ± 0.07 mm. The average value of the pectinolytic index in the isolate UNJCC Y-144 was not significantly different from the pectinolytic index in the isolate UNJCC Y-149 with a value of 0.49 ± 0.06 mm. This indicated that UNJCC Y-144 and UNJCC Y-149 yeasts had the same pectinolytic index value. Obtained isolates from the yeast Debaryomyces hansenii and Candida zeylanoides having pectinolytic index values of 4.3 mm and 5.3 mm, respectively.27 The identification results based on the D1/D2 rDNA region of yeast isolates UNJCC Y-144, UNJCC Y-148, and UNJCC Y-149 are known to belong to the order Saccharomycetales is an order of the class Saccharomycetes and is an ascomycetes yeast that has an ascus formed by the fusion of gametangia from adjacent cells.48 With a bootstrap score of 99%, isolate UNJCC Y-144 belongs to a monophyletic group that includes Cy. fabianii. With a 95% bootstrap value, isolates UNJCC Y-148 and UNJCC Y-149 belonged to the same monophyletic group as yeast C. tropicalis. A clade is reliable if its bootstrap value is close to 90%.49
Aspergillus welwitschiae was the outgroup that was utilised to build the phylogenetic tree using the D1/D2 rDNA region. Outgroups are required to supply features or characters in the form of plesiomorphic and apomorphic characters.50 Plesiomorphic characters are distinct primitive characters that are found in the outgroup, whereas apomorphic characteristics are discovered and inherited in the ingroup.51 Outgroups are not included in the group despite having a strong correlation with species groups.52
Relative enzyme activity was calculated for the three yeast isolates, Cy. fabianii UNJCC Y-144, C. tropicalis UNJCC Y-148, and C. tropicalis UNJCC Y-149 using carbon and nitrogen sources variations. The selection of variations of carbon sources and nitrogen sources aims to determine which yeast, type of carbon source, and nitrogen source are optimal in producing pectinase enzymes. To get crude enzyme extract, the fermentation procedure was conducted utilising the Solid-state fermentation (SSF) technique. Using a no soluble substance that serves as both a source of nutrients and a solid support, SSF is carried out in the absence of a free-flowing liquid that contains enough water to sustain the development of microorganisms. This technique is employed because to its simplicity of preparation and adaptability to changes in temperature, pH, and oxygen concentration in liquid media.53 The crude extract of the enzyme was obtained by extracting the fermentation medium after 48 h incubation. Suggests that 48 h is the optimum time to produce pectinase enzymes.27 Used an incubation time of 48 h because it is the optimum time for pectinase production in a liquid medium.54 The fermentation medium was agitated on a rotator shaker at a speed of 150 rpm for 48 h. The purpose is to mix and transfer oxygen, which affects the enzyme formation process.55 Dissolved oxygen in the medium produced in the agitation process affects the secretion of pectinase enzyme activity.56 After 48 h, 5 mL of the filtrate was taken and centrifuged at 3000 rpm for 9 min at 4 °C. Centrifugation at 4 °C will not cause enzyme degradation.57 Centrifugation was done to obtain a crude extract supernatant of the enzyme and separate the enzyme from other particles.
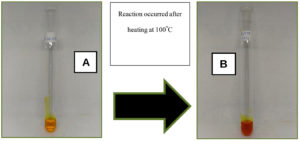
Based DNS approach, the polygalacturonase enzyme activity was quantitatively tested.32 The formula for the pectinase enzyme’s activity was then applied to the acquired absorption value. Three-way ANOVA was used to analyse the acquired enzyme activity value (U/mL) and determine the impact and interplay of yeast isolate variables, carbon source type, and nitrogen source type on pectinase enzyme activity. That carbon and nitrogen supplies are critical to the fermentation medium because yeast uses them for cell development and other metabolic activities that impact the synthesis of pectinase enzymes.36 A standard glucose curve must be created in order to calibrate and determine the value of the pectinase enzyme activity.33 With a correlation value (R2) of 0.9872, the standard curve is utilised as the linear equation y = 0.4514x + 0.0228. The correlation value (R2), which is near to 1, suggests that there is a link between the absorbance value and glucose content.58 The DNS approach was used to do a quantitative assay of relative enzyme activity. This method aims to measure the activity of the pectinase enzyme based on the reducing sugar content resulting from the hydrolysis of the pectinase enzyme to the substrate.59 The principle of testing using the DNS method is an oxidation-reduction reaction that can change the yellow color to reddish orange (Figure 9).
Figure 9. Reduction reaction of 3,5-dinitrosalicylic acid (DNS) to 3-amino-5-nitrosalicylic (A) DNS before being heated at 100 °C for 15 minutes; (B) The reaction that occurs after heating
According to Toledo et al. the reaction that occurred was the reduction of 3,5-dinitrosalicylic acid (DNS) to 3-amino-5-nitrosalicylic acid (Figure 9). The aldehyde group of the sugar is reduced to a carboxyl group. DNS as an oxidizing agent will be reduced to form 3-amino-5-nitrosalicylic acid. States that the more concentrated the color formed, the more it is assumed that, the more reducing sugar content contained in the sample, and the higher the absorbance value produced.60
Based on the results obtained, Cy. fabianii UNJCC Y-144 has a maximum relative enzyme activity value in a medium with sucrose and urea. While it is shallow in the medium containing pectin, isolate Cy. fabianii UNJCC Y-144 showed noticeably high enzyme activity in the media without pectin as a substrate molecule. It is anticipated that pectin is essential for Cy. fabianii UNJCC Y-144 to produce pectinase because it acts as an inhibitor. Other chemicals have the ability to control the activity of enzymes, making them more or less active. Enzyme activators are compounds that make an enzyme more active, whereas enzyme inhibitors are substances that make an enzyme less active. Cy. Fabianii, which was previously Candida fabianii, Hansenula fabianii, and Pichia fabianii, is an ascomycetes with a white to cream colony and round cell shape.61-63 This strain can be obtained from fruits such as olives and sugarcane and has been used to treat waste from food processing.64
Isolate C. tropicalis UNJCC Y-148 and C. tropicalis UNJCC Y-149 exhibited low pectinase activity in almost all mediums. It indicates that the intake of CN source by the two isolated is not done optimally to produce pectinase. It can also be assumed that with the current concentration of CN used, it contributes only a little to the production of pectinase enzyme by C. tropicalis UNJCC Y-148 and C. tropicalis UNJCC Y-149. C. tropicalis is a yeast with a fine colony texture and round cell shape, and a size ranging from 2-5 x 3-7 m.34 C. tropicalis can be isolated from various fruit substrates, including citrus fruits and mangaba.42 According to research by Pauldas & Jain, C. tropicalis can produce pectinase enzymes at the optimum temperature of 35 °C, marked by its ability to degrade pectin substances in the fruit juice clarification process.
The highest enzyme activity value of C. tropicalis UNJCC Y-148 was only 10.63 ± 0.97 U/mL, obtained in the medium with glucose carbon source and nitrogen peptone source. The explanation is that glucose and peptone are predicted to play together as activators of the pectinase enzyme in C. tropicalis UNJCC Y-148. Pectin might also play as an activator, but with the availability of specific molecules such as peptone and urea as nitrogen sources. In the case of C. tropicalis UNJCC Y-149, the highest value of enzyme activity obtained was only 4.41 ± 0.64 U/mL in a medium with glucose as a carbon source and peptone as a nitrogen source. The level of enzyme activity in the medium containing pectin was slightly higher; it can be thought that pectin plays a role as an activator molecule with the availability of other molecules in pectinase production by C. tropicalis UNJCC Y-149. With the optimum concentration of CN ratio in the medium, a higher level of pectinase activity might be reached.
Based on the results of S. cerevisiae UNJCC Y-87, the highest pectinase enzyme activity was obtained in the medium with a carbon source of sucrose and a nitrogen source of urea with an activity value of 35.03 ± 5.62 U/mL. The lowest activity value was obtained in a medium with carbon sucrose and nitrogen source ammonium chloride, with a pectinase enzyme activity value of 0.57 ± 0.14 U/mL. S. cerevisiae UNJCC Y-87 presented a higher level of pectinase activity in the medium containing peptone with the addition of simpler carbon such as glucose or sucrose and urea with the addition of sucrose. Medium containing ammonium chloride as a nitrogen source or pectin as carbon shows very low enzyme activity. It can be assumed that the two molecules play as inhibitors for pectinase production in S. cerevisiae UNJCC Y-87. According to Enshasy et al., using a carbon source of sucrose in the fermentation process can increase the activity of the pectinase enzyme with an enzyme activity value of 450 U/mL. Sucrose is a disaccharide which is broken down by yeast into glucose and fructose.36
Five isolates out of 20 representative yeasts were collected from UNJCC, which showed pectinase enzyme activity after semi-quantitative screening and quantitative screening with the DNS method. The isolate codes of UNJCC Y-144, UNJCC Y-148, UNJCCY-149, UNJCC Y-150, and UNJCC Y-151 were obtained. The D1/D2 rDNA regional sequence analysis results showed that the isolate UNJCC Y-144 has a homology level of 100% with Cyberlindnera fabianii with a bootstrap value of 99%. Isolate UNJCC Y-148 and UNJCC Y-149 have homology levels of 100% and 99.83%, respectively, with Candida tropicalis with a bootstrap value of 95%. This study shows that isolate types, carbon and nitrogen sources, and environmental conditions affected the yeast isolates’ secretion of pectinase. The highest enzyme activity value was 48.68 ± 2.84 U/mL, obtained from isolated Cyberlindnera fabianii UNJCC Y-144 in a medium containing a carbon source of sucrose and a nitrogen source of urea. It is necessary to optimize the ratio of carbon and nitrogen sources, incubation time, temperature, pH, and the amount of inoculum that can affect the activity of the pectinase enzyme.
ACKNOWLEDGMENTS
This study was supported by funding from the National Research and Innovation Agency of Indonesia through Program Grant No. 12/II.7/HK/2023-2024, awarded to Dalia Sukmawati. The project, titled “Alternative Nutritious Food Security Based on Black Soldier Fly (Hermetia illucens) and Probiotic Oleaginous Yeast Through a Metabolomic Approach,” focuses on creating sustainable food security solutions.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
This study was funded by the National Research and Innovation Agency of Indonesia through Program Grant No. 12/II.7/HK/2023-2024.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- Mohnen D. Pectin structure and biosynthesis. Curr Opin Plant Biol. 2008;11(3):266-277.
Crossref - Gummadi SN, Panda T. Purification and biochemical properties of microbial pectinases – A review. Process Biochem. 2003;38(7):987-996.
Crossref - Rashad MM, Abdou HM, Shousha WGH, Ali MM, El-Sayed NN. Purification and characterization of the pectin lyase produced by Pleurotus ostreatus grown on lemon pulp waste. Aust J Basic Appl Sci. 2011;5(8):1377-1384.
- Swain MR, Ray RC. Production, Characterization and application of a thermostable exo- polygalacturonase by Bacillus subtilis CM5. Food Biotechnol. 2010;24(1):37-50.
Crossref - Namasivayam E, John RD, Mariappan K, Jiji A, Kumar M, Richard L. Production of extracellular pectinase by Bacillus Cereus isolated from market solid waste.
J Bioanal Biomed. 2011;3(3):70-75.
Crossref - Amin F, Bhatti HN, Bilal M. Recent advances in the production strategies of microbial pectinases-A review. Int J Biol Macromol. 2019;122:1017-1026.
Crossref - Anggraini DP, Roosdiana A, Prasetyawan A, Mardiana Dx. Effect of Metal Ions on Pectinase Activity from Aspergillus niger on Purifying Guava Fruit. Natural B. 2002;2(1):66-72.
- Phutela U, Dhuna V, Sandhu S, Chadha BS. Pectinase and polygalacturonase production by a thermophilic Aspergillus fumigatus isolated from decomposting orange peels. Braz J Microbiol. 2005;36(1).
Crossref - Garg G, Singh A, Kaur A, Singh R, Kaur J, Mahajan R. Microbial pectinases: an ecofriendly tool of nature for industries. 3 Biotech. 2016;6(1):47.
Crossref - Sivaramakrishnan S, Gangadharan D, Nampoothiri KM, Soccol CR, Pandey A. a-Amylases from microbial sources – an overview on recent developments. Food Technol Biotechnol. 2006;44(2):173-184.
- Angayarkanni J, Palaniswamy M, Murugesan S, Swaminathan K. Improvement of tea leaves fermentation with Aspergillus spp. pectinase. J Biosci Bioeng. 2002;94(4):299-303.
Crossref - Jayani RS, Saxena S, Gupta R. Microbial pectinolytic enzymes: A review. Process Biochem. 2005;40(9):2931- 2944.
Crossref - Sukmawati D, Sunari A, Akbari TS, et al. Isolation and selection of fungi amylolytic and cellulolytic originated from Rhododendron sp. flower. AIP Conf Proc. 2020;2242(1):50023.
Crossref - Sukmawati Dalia, Arman Z, Sondana GA, et al. Potential amylase-producing yeast isolated from indigenous fermented beverages originating from Bali, Indonesia potential amylase-producing yeast isolated from indigenous fermented beverages originating from Bali, Indonesia. J Phys Conf Ser. 2019;1402(5):055021.
Crossref - Risandi A, Fuady R, Sukmawati D, et al. Isolation and screening of amylolytic yeast from Paphiopedilum sp, originating from Bedugul Botanical Garden, Bali, Indonesia isolation and screening of amylolytic yeast from Paphiopedilum sp, originating from Bedugul Botanical Garden, Bali, Indonesia. J Phys Conf Ser. 2019;1402(3):140233060.
Crossref - De Becze GI. A microbiological process report; yeasts. I. Morphology. Appl Environ Microbiol. 1955;4(1):1-12.
Crossref - Alimardani-theuil P, Gainvors-claisse A, Duchiron F. Yeasts: An attractive source of pectinases – from gene expression to potential applications: A review. Process Biochem. 2011;46(8):1525-1537.
Crossref - Ardhana MM, Fleet GH. The microbial ecology of cocoa fermentations in Indonesia. Int J Food Microbiol. 2003;86(1):87-99.
Crossref - de Oliveira APA, Silvestre MA, Alves-Prado HF, et al. Bioprospecting of yeasts for amylase production in solid state fermentation and evaluation of the catalytic properties of enzymatic extracts. Afr J Biotechnol. 2015;14(14):1215-1223.
Crossref - Dellanerra D, Risandi A, Sunari A, Sukmawati D, Husna SNA, El-Enshasy HA. Screening and characterization of amylolitic mold originated from ghost crab (Ocypode sp.) in Cidaon, Ujung Kulon National Park, Indonesia. In AIP Conf Proc. 2019;2120(1).
- Belgis M, Wijaya CH, Apriyantono A, Kusbiantoro B, Yuliana ND. Volatiles and aroma characterization of several lai (Durio kutejensis) and durian (Durio zibethinus) cultivars grown in Indonesia. Scientia Horticulturae. 2017;220:291-298.
Crossref - Picot-Allain MCN, Ramasawmy B, Emmambux MN. Extraction, characterisation, and application of pectin from tropical and sub-tropical fruits: a review. Food Rev Int. 2022;38(3).
Crossref - Hariyati T, Kusnadi J, Arumingtyas EL. Genetic diversity of hybrid durian resulted from cross breeding between Durio kutejensis and Durio zibethinus based on random amplified polymorphic DNAs (RAPDs). Am J Mol Biol. 2013;3(3):153-157.
Crossref - Alagarsamy S, Larroche C, Pandey A. Microbiology and industrial biotechnology of food-grade proteases: a perspective. Food Technol Biotechnol. 2006;44(2):211.
- Blanco P, Sieiro C, Villa TG. Production of pectic enzymes in yeasts. FEMS Microbiol Lett. 1999;175(1):1- 9.
Crossref - Oskay M, Yalcin HT. Screening of yeast strains for pectinolytic activity: effects of different carbon and nitrogen sources in submerged fermentations. OnLine J Biol Sci. 2015;15(3):89-96.
Crossref - Sukmawati D, Setyaningsih A, Handayani KT, Rahayu S, Rustam Y, Moersilah M, Husna SNA. Isolation and characterization of aflatoxigenic Aspergillus spp. from maize of livestock feed from Bogor. In IOP conf ser: Mater Sci Eng. 2018;434:012105.
- Sukmawati Dalia, Nurkhasanah S, Afifah ZN, et al. Metagenomic-based approach for the analysis of yeast diversity associated with amylase production in lai (Durio Kutejensis). J Pure Appl Microbiol. 2021;15(1):75-90.
Crossref - Makene VA. Identification of non-albicans Candida yeasts associated with vulvovaginal candidiasis in Tanzania using a combination of multiplex PCR and DNA sequence divergence of the 26S LSU rDNA. Scholars Acad J Biosci. 2014;2(2):124-131.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725-2729.
Crossref - Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem. 1959;31(3):426-428.
Crossref - Takci HAM, Turkmen FU. Extracellular pectinase production and purification from a newly isolated Bacillus subtilis strain. Int J Food Prop. 2016;19(11).
Crossref - Silva S, Negri M, Henriques M, Oliveira R, Williams DW, Azeredo J. Candida glabrata, Candida parapsilosis and Candida tropicalis: biology, epidemiology, pathogenicity and antifungal resistance. FEMS Microbiol Rev. 2012;36(2):288-305.
Crossref - Deorukhkar SC, Roushani S. Identification of Candida species: conventional methods in the era of molecular diagnosis. Ann Microbiol Immunol. 2018;1(1):1002.
- El Enshasy HA, Elsayed EA, Suhaimi N, Malek RA, Esawy M. Bioprocess optimization for pectinase production using Aspergillus niger in a submerged cultivation system. BMC Biotechnol. 2018;18(1):71.
Crossref - Maidana SA, Esteche VP, Hours RA, Brumovsky LA, Martos MA. Use of agro-industrial wastes for the production of a wild yeast enzyme with disintegration activity on plant tissues. Braz Arch Biol Technol. 2020;63:1-13.
Crossref - Patil RSR, Dayanand A. Production of pectinase from deseeded sunflower head by Aspergillus niger in submerged and solid-state conditions. Bioresour Technol. 2006;97(16):2054-2058.
Crossref - Purwadaria T, Nirwana N, Ketaren PP, Iswantini PD, Widyastuti Y. Synergistic activity of enzymes produced by Eupenicillium javanicum and Aspergillus niger nrrl 337 on palm oil factory wastes. Biotropia. 2003;20.
Crossref - Haile M, Kang WH. Isolation, identification, and characterization of pectinolytic yeasts for starter culture in coffee fermentation. Microorganisms. 2019;7(10):401.
Crossref - Martos MM, Zubreski ER, Combina M, Garro OA, Hours RA. Isolation of a yeast strain able to produce a polygalacturonase with maceration activity of cassava roots. Food Sci Technol. 2013;33(2):332-338.
Crossref - Silva D, Tokuioshi K, Martins ES, Da Silva R, Gomes E. Production of pectinase by solid-state fermentation with Penicillium viridicatum RFC 3. Process Biochem. 2005;40(8):2885-2889.
Crossref - Kusyanto K, Rachmadina S. The Effect of H2SO4 Concentration and Micro Wave Power in Microwave Assisted Hydrolysis of Furfural Production from Empty Palm Fruit Bunches. Jurnal Bahan Alam Terbarukan. 2015;4(1):14-20.
Crossref - Kashyap DR, Vohra PK, Chopra S, Tewari R. Applications of pectinases in the commercial sector: a review. Bioresour Technol. 2001;77(3):215-227.
Crossref - Minotto E, Milagre LP, Oliveira MT, Van Der Sand ST. Enzyme characterization of endophytic actinobacteria isolated from tomato plants. J Adv Sci Res. 2014;5(02):16-23.
- Valgas C, de Souza SM, Smania EFA, Jr AS. Screening methods to determine antibacterial activity of natural products. Braz J Microbiol. 2007;38(2):369-380.
Crossref - Sudeep KC, Upadhyaya J, Joshi DR, et al. Production, characterization, and industrial application of pectinase enzyme isolated from fungal strains. Fermentation. 2020;6(2):59.
Crossref - Suh S-O, Blackwell M, Kurtzman CP, Lachance M-A. Phylogenetics of Saccharomycetales, the ascomycete yeasts. Mycologia. 2006;98(6):1006-1017.
Crossref - Symonds MRE, Elgar MA. Phylogeny affects estimation of metabolic scaling in mammals. Evolution. 2002;56(11):2330-2333.
Crossref - Hidayat T, Pancoro A. Molecular Phylogenetic Studies and Their Role in Providing Basic Information to Improve the Quality of Orchid Genetic Resources. AgroBiogen Journal. 2018;4:35-40.
Crossref - Pangestika Y, Budiharjo A, Kusumaningrum HP. Phylogenetic Analysis of Curcuma Zedoaria (White Temu) Based on Internal Transcribed Spacer (ITS) Gene. Jurnal Akademika Biologi. 2015;4(4)8-13.
- Sukmawati D, Andrianto MH, Arman Z, Ratnaningtyas NI, Al Husna SN, El-Enshasy HA, Kenawy AA. Antagonistic activity of phylloplane yeasts from Moringa oleifera Lam. leaves against Aspergillus flavus UNJCC F-30 from chicken feed. Indian Phytopathology, 2020;73:79-88.
- Ravichandran S, Vimala R. Solid state and submerged fermentation for the production of bioactive substances: a comparative study. Int J Sci Nat. 2012;3:480-486.
- Nath M, Majumdar B, Das S, Mazumdar SP, Saha AR, Sarkar S. Optimization of fermentation conditions for pectin degrading enzyme production by pectinolytic microbial consortia used for jute retting. Int J Curr Microbiol Appl Sci. 2017;6(11):925-931.
Crossref - Gupta P, Gigras P, Mohapatra H, Goswami VK, Chauhan B. Microbial a-amylases: a biotechnological perspective. Process Biochem. 2003;38(11):1599- 1616.
Crossref - Ibrahim D, Weloosamy H, Lim S-H. Effect of agitation speed on the morphology of Aspergillus niger HFD5A-1 hyphae and its pectinase production in submerged fermentation. World J Biol Chem. 2015;6(3):265-271.
Crossref - Huyet J, Ozeir M, Burgevin MC, et al. Structural insights into the forward and reverse enzymatic reactions in human adenine phosphoribosyltransferase. Cell Chem Biol. 2018;25(6):666-676.e4.
Crossref - Reddy MPC, Saritha KV. Effects of the culture media optimization on pectinase production by Enterobacter sp. PSTB-1. 3 Biotech. 2016;6.
Crossref - Haile S, Masi C, Tafesse M. Isolation and characterization of pectinase-producing bacteria (Serratia marcescens) from avocado peel waste for juice clarification. BMC Microbiol.2022;22:145.
Crossref - Solihin J, Waturangi DE, Purwadaria T. Induction of amylase and protease as antibiofilm agents by starch, casein, and yeast extract in Arthrobacter sp. CW01. BMC Microbiol. 2021;21:232.
Crossref - Bauer R, Begerow D, Oberwinkler F, Marvanova L. Classicula: the teleomorph of Naiadella fluitans. Mycologia. 2003;95(4):756-764.
Crossref - Kurtzman CP, Robnett CJ. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Anton Leeuwen. 1998;73:331-371.
- Park J-H, Oh J, Sang H, et al. Identification and antifungal susceptibility profiles of Cyberlindnera fabianii in Korea. Mycobiology. 2019;47(4):1-8.
Crossref - Freel KC, Sarilar V, Neuveglise C, Devillers H, Friedrich A, Schacherer J. Genome sequence of the yeast Cyberlindnera fabianii (Hansenula fabianii). Genome Announcements. 2014;2(4).
Crossref - Pauldas K, Jain A. Optimization of pectinase production kinetics by Candida tropicalis and its applications in fruit juice clarification. Int J Pharm Biol Sci. 2018;8(3):946-958.
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.