ISSN: 0973-7510
E-ISSN: 2581-690X
Biosurfactants were found to be a suitable candidate for environment clean-up and as an alternative to conventional treatment systems. In the present study, a polyaromatic hydrocarbon degrading organism Bacillus halotolerans was screened for its ability to produce biosurfactant during hydrocarbon degradation. The bio-surfactant activity of the organism was screened by using various methods like oil spreading technique, emulsification assay, foam height analysis and parafilm-M test. The design expert software was used to optimize the suitable conditions for the production of biosurfactants. The optimum conditions were determined as pH 6, Chrysene-100 ppm, nitrogen source -1000 ppm and at 144 hrs using the design of experiments. The organism showed good oil degradation capacity and maximum growth was achieved in 6 days. Biosurfactant obtained from the biodegradation medium was confirmed to be lipopeptide using TLC, FTIR and GCMS.
Bacillus halotolerans, Biosurfactant, Lipopeptides, Bioremediation, Hydrocarbons
Surfactants are molecules that modify the interfacial properties between two fluids, or that between a fluid and a solid. They contain both hydrophobic and hydrophilic moieties.1 It serves two essential jobs: First, it assists with further developing contact with the surface. Furthermore, it emulsifies oils and fats. Surfactants can be either oil or vegetable-based. The drawbacks of utilizing oil-based surfactants are that they are produced from a non-renewable source, slightly biodegradable, and discharge poisonous synthetic compounds when they break down. The fundamental benefit of utilizing vegetable-based surfactants is that they are obtained from sustainable sources.2 Surfactant molecules which are produced by microorganisms are referred as biosurfactants and most of the hydrocarbon degraders are found to be biosurfactant producers.3 Biosurfactant exhibit lower toxicity than the synthetic surfactants.4 Based on the microbial source and chemical composition, the biosurfactants are classified into four groups: they are lipopeptides, phospholipid, polymeric surfactants and glycolipids.5,6 Glycolipids are carbohydrates linked to lipids by glycosidic bond. Examples include trehalose lipids, rhamnolipids and sophorolipids.7 These are produced by organisms like Pseudomonas spp.8,9 Torulopsis spp.10,11 Lipopeptides are molecules containing lipids linked to short peptide chains whereas in lipopolysaccharides, lipids are attached to polysaccharides. Lipopeptides include Surfactin, Lichenysin, Iturin and Fengycin. Most of these lipopeptides are produced by Bacillus subtilis.6,12 Lipopolysaccharides, for example, emulsan, is produced by Acinetobacter spp.12 Polymeric surfactants like nonionic trehalose corynomycolates are produced by Rhodococcus, Arthrobacter spp. and various Mycobacterium spp.13 Among fungi, Candida bombicola,14 Candida lipolytica,15 Candida ishiwadae,16 Candida batista,17 Aspergillus ustus18 and Trichosporon ashii19 are the explored ones. Even though glycolipids are the major type produced, lipopeptides are the most powerful ones.20
Biosurfactants allow the microbes to grow on water-immiscible substrates by lowering the substrate tension, thus making the substrates readily available for the uptake and metabolism. Other physiological roles include antimicrobial activity and cell adherence.4 Biosurfactants have the properties of frothing, wetting, dropping surface tension, settling emulsions and are normally non-toxic and biodegradable.21 Their applications include cosmetics, pharmaceuticals, humectants, petroleum extraction and refining, food additives and detergents. The composition and the production of biosurfactants depends on strain, culture conditions like carbon source, nitrogen source, chemical and physiological parameters such as temperature, pH, aeration, divalent cations etc.22 In spite of all laboratory-based success in the production of biosurfactants, industry level is still challenging because of many factors like medium and concentration of biosurfactants. In the present study, we aim to screen the biosurfactant producing capability of the strain isolated from oil contaminated sites and to optimize the production process using RSM. Our focus is to use Chrysene, a polyaromatic hydrocarbon as the major carbon source for the production, as there is only limited statistics about the compound.
Bacteria and Culture Condition
In the present study, Bacillus halotolerans (Genbank Accession number MK439524) which was isolated from oil contaminated water samples was selected for the production of the biosurfactant.23 A 24 hour grown culture was used as the inoculum for the production process.
Chemicals and Media
All the chemicals used in the study were purchased from Himedia, except Chrysene (Sigma) with a greater than 98.0% analytical standard.
Screening for the Biosurfactant Production
Minimal salt broth supplemented with 1% chrysene and 1 ml crude oil at a pH of 7.0 was used for the initial screening. To that 5 ml of 24-hour old culture was added and incubated at 30°C in an orbital shaker (150 rpm) for 5 days. The samples were tried for the capacity to deliver of biosurfactants by performing following tests:
Oil Spreading Technique
Oil spreading experiment was performed as described by Morikawa et al.24 In this method, 10μl of kerosene oil was added into a petri dish containing 50 ml of distilled water. 10μl of the culture filtrate was then dropped onto the middle of the oil layer. Formation of a clear zone indicates the presence of biosurfactants. The diameter of the clear zone is measured after 30 seconds.
Emulsification Assay
Emulsification activity was observed by adding equal amounts of kerosene oil and cell free supernatant. It was then vortexed for 2 min and was left undisturbed for 24 hours. Later the emulsification activity was measured using the following equation3:
EI = (Emulsion height)/ (Total height) ×100
Foam Height Analysis
Foam height was determined by vortexing 10mL of the cell free supernatant for 10 min and left to stand for 2 minutes. Foam height was measured and frothing capacity was measured using the equation25:
Foaming = (Foam height)/ (Total height) ×100
Effect of Media on Growth and Biosurfactant Production
A statistical approach, response surface method was employed to optimize various parameters26 for ensuring better production of the surfactant. The parameters selected for the study were carbon source, incubation period(days), pH, and concentration of nitrogen sources. The range for the initial optimization factor taken as Table 1. After initial optimization, higher and lower levels were taken and those values given to design expert software. The experiment was designed and conducted as instructed by software, Design of Experiments. Four variable arithmetical components were inspected at two usually separated levels, picked like +1 (high level) and -1 (low level) in triplicates.
Table (1):
The range of process parameters for the Design of Experiments.
Term |
Parameter |
Low Level |
High Level |
|---|---|---|---|
A |
No.of days |
72 |
192 |
B |
pH |
6 |
8 |
C |
Carbon source (mg/l) |
50 |
100 |
D |
Nitrogen source (mg/l) |
500 |
2000 |
Extraction and Characterization of Biosurfactant Produced
For the extraction of biosurfactant, inoculum was added into minimal salt media optimized as per the statistical method. At the end of the incubation period, bacterial pellets were removed by centrifugation at 10000 rpm for 20 min at 4°C (REMI C-24BL cooling centrifuge). Concentrated sulphuric acid was added to the resulting supernatant and kept at 4°C overnight. The centrifugation was repeated and a Grey white precipitate was obtained. Further extraction was done with 10 mL of chloroform methanol (2: 1 v/v) and the content was centrifuged at 10000 rpm for 20 min under cooling condition and the resulting pellet was air dried.27 The extracted biosurfactant was characterized using thin layer chromatography, FTIR and GC–MS.
Thin Layer Chromatography
The crude sample was spotted into silica gel 60 F254 TLC plates and developed using chloroform: methanol: acetic acid (65:15:1) as a solvent system. Type of the biosurfactant was visualized using various agents, such as 0.2% ninhydrin in ethanol for lipopeptide, 1% sulphuric acid followed by heating at 110°C for 20 min for glycolipids and iodine vapours for lipids. The Rf value was calculated using the below formula.28
Rf = Distance travelled by the sample /Distance travelled by the solvent front.
Parafilm-M Test
Two ml of the culture filtrate was mixed with a few drops of Bromophenol blue indicator. 10 μl of the sample was dropped onto a parafilm-sheet with a micropipette and the shape of the drop was inspected after 1 min. Phosphate buffer of pH 7.0 and Sodium lauryl sulfate and were used as positive and negative controls respectively.29
FTIR and GC–MS
The crude biosurfactant sample was subjected to infrared analysis to identify its chemical nature.29 The analysis was carried out using Shimadzu IR prestige-21 FTIR Spectrophotometer. The spectral data was collected within a range of 600 to 4000 cm-1. The obtained spectra were further analysed for the presence of various functional groups and chemical interactions. The methanol extract of the sample was used for the GC-MS analysis.30 The sample is injected into the instrument (Shimadzu QP2010S), which was equipped with a Rxi-5Sil MS column (30 m × 0.25 mm).
Biodegradation of Crude Oil
Spectrophotometric method was used to assess the efficiency of the organism to degrade crude oil. Bacterial cells were inoculated into the minimal salt medium (MSM) containing 7% crude oil as a sole carbon source. The uninoculated medium serves as the negative control. All the samples were incubated in an orbital shaker at 180 rpm and 28°C for 8 days. To determine the reduction in the amount of oil, the absorbance of the sample was measured at 610 nm in every 24 hours.31 The sample composition was determined using GC-MS.
Screening for the Biosurfactant Production
The organism was screened for the production of biosurfactant and the results were depicted as given below:
Oil Spreading Technique
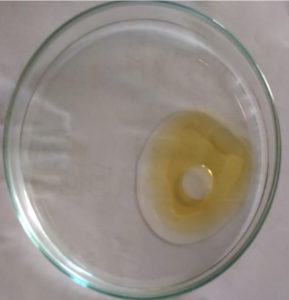
Cell free sample was dropped carefully on to the middle of the oil layer taken in the Petri plate. The organism was able to displace the oil and presented a clear zone (Figure 1). Diameter of the oil drop was 9mm and later it widened to a diameter of 1.3cm. Distilled water and a synthetic surfactant Triton X-100 was used as negative and positive controls respectively.32 Appearance of a clear zone confirmed the presence of biosurfactants.
Emulsification Assay
Production of biosurfactant is also confirmed by determining the emulsification index and it was found to be 41.05 (Figure 2). The value is slightly lower in comparison with biosurfactants from Bacillus spp.33 This might be due to the difference in the carbon source used for the production. The ability of the organism to produce biosurfactant was confirmed.
Foaming Activity
Foaming ability of crude biosurfactant was found to be more than 50% and stable for 1 hour (Figure 3). The foam stability was observed continuously at a 5 min interval for 1 hour and after 1 hour, there was a reduction in the foam height by 2%.
Optimization of Biosurfactant Production
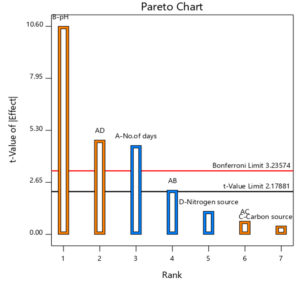
Using the design expert software (version 11), various experimental combinations were found out and performed. The observations were recorded as OD at 610 nm. Using this, the most suitable condition of the media for the production of biosurfactant was determined. All the process parameters were studied and optimized using RSM. The results of ANOVA for the factorial model were shown in Table 2. The model is found to be relevant as the F- value is 23.25. There is just a 0.01% possibility that this large F-value could happen because of noise. P< 0.05 demonstrates that the factors such as No. of days(A), pH(B), combinations such as No. of days and pH (AB) and No. of days and nitrogen source (AD) are relevant models. The fit analysis shows a value of 0.7547 and 0.8964 for predicted and adjusted R2 respectively. As the differences between both the terms are negligible, the model is significant. The signal to noise ratio was found to be 14.345, indicating adequate signal and hence the present model can be used to explore the design space.
Table (2):
ANOVA analysis.
Factors |
SS |
df |
Mean Sq. |
F-value |
p-value |
|---|---|---|---|---|---|
Model |
4.91 |
7 |
0.7015 |
23.25 |
< 0.0001 |
No.of days (A) |
0.6170 |
1 |
0.6170 |
20.45 |
0.0009 |
pH(B) |
3.39 |
1 |
3.39 |
112.27 |
< 0.0001 |
Carbon source(C) |
0.0054 |
1 |
0.0054 |
0.1790 |
0.6804 |
Nitrogen source(D) |
0.0414 |
1 |
0.0414 |
1.37 |
0.2661 |
AB |
0.1521 |
1 |
0.1521 |
5.04 |
0.0463 |
AC |
0.0135 |
1 |
0.0135 |
0.4460 |
0.5180 |
AD |
0.6939 |
1 |
0.6939 |
23.00 |
0.0006 |
Curvature |
0.7912 |
1 |
0.7912 |
26.22 |
0.0003 |
Pure Error |
0.3319 |
11 |
0.0302 |
||
Cor Total |
6.03 |
19 |
The coefficient estimates address only a normal change in factor esteem when all leftover variables are held steady. The catch in a symmetrical plan is the general normal reaction of the relative multitude of runs. At the point when the components are symmetrical the VIFs are 1; VIFs more noteworthy than 1 demonstrate multi-collinearity, the higher the VIF the more extreme the connection of variables. VIFs less than 10 are acceptable in most of the cases. The concluding equation in terms of coded factors is represented as:
R1=0.7005-0.1964A+0.4601B+0.0184C-0.0509D-0.0975AB+0.0290 AC+0.2082AD
The equation based on coded components can be utilized to make expectations about the reaction for limits of each component. Naturally, the undeniable limits of the components are coded as +1 and the lower limits are coded as -1. The coded condition is helpful for distinguishing the overall effect of the variables by looking at the factor coefficients. The final equation with respect to the real factors can be utilized to foresee the reaction of given levels of each factor and is represented as:
R1=0.000867A+0.674625B-0.001817C-0.000679D-0.001625AB+0.000019AC+4.62778E-06-2.60520
where A-number of days, B-pH, C- chrysene, D- nitrogen source.
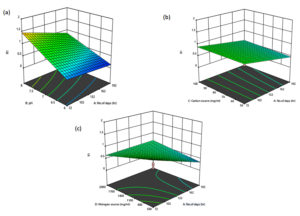
Various factors such as Carbon source, combination of no. of days and nitrogen source, no. of days and carbon source shows a positive effect as shown in the Figure 4 and 5. By using the point prediction, the maximum growth of the organism was obtained at pH 8, carbon source -75mg/ml, nitrogen source -1250mg/ml and at 72 hrs.
Figure 4. Interaction between (a) No. of days and pH (b) Carbon source and No. of days (c) Nitrogen source and No. of days
Extraction and Characterization
The cell free extract was treated with concentrated sulphuric acid followed by chloroform methanol. The resultant precipitate was air dried and preserved for screening (Figure 6).
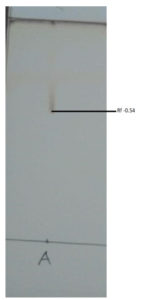
The TLC plates with the sample after treatment with ninhydrin produced a red spot as shown in the Figure 7. This confirmed the presence of lipopeptides in the extracted biosurfactant. The Rf value was found to be 0.54. Most of the Bacillus spp., produce lipopeptides belonging to the surfactin family. Identical results were reported by Joy et al.34 on Bacillus spp. (SB2) which was confirmed on TLC.
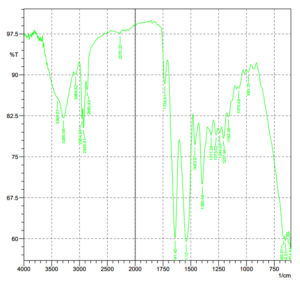
The IR spectra of the biosurfactant sample extracted from Bacillus halotolerans is shown in Figure 8 and compared with previously reported works. The spectrum displays various peaks corresponding to different functional groups and chemical bonds (Table 3).
Table (3):
Signature peaks of lipopeptides.
No |
Peaks |
Functional groups |
|---|---|---|
1 |
3290 cm-1 |
Peptide (O=C-NH)29 |
2 |
2926 cm-1 |
Aliphatic chain (H-C=C-H)35 |
3 |
1734 cm-1 |
Esters (RCO2R’)36 |
4 |
1541 cm-1 |
Carbonyl group (C=O)37 |
The data is in good correlation with the lipopeptide nature of the biosurfactant. Hence it can be confirmed that the extracted biosurfactant has a similar structure and functional group to lipopeptides.
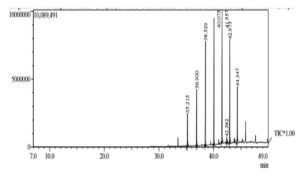
The biomolecule was further analysed for its fatty acid composition using GC-MS and revealed that the biosurfactant extracted from B. halotolerans was found to be lipopeptide in nature (Figure 9). Major peaks were Octacosane (C28H58), Nonadecane (C19H40), and Eicosane (C20H42). Other components obtained were Tetracosane (C24H50), Heneicosane (C21H44), Tricosane (C23H48), Nonadecane, 2-methyl (C20H42). Qiao and Shao30 reported the presence of decanoic compounds in lipopeptide surfactants. The composition of the lipopeptide molecule depends not only on the organism, but is affected by the substrates and culture conditions. It was also reported that the bioactive molecule produced by B. subtilis was reported to be lipopeptide in nature.38 Our earlier studies could confirm that Bacillus halotolerans is closely related to B. subtilis.23 Further analysis is required to identify the type of lipopeptide surfactant.
Parafilm-M Test
In the parafilm M test, the drop became flat for sample and sodium lauryl sulphate (positive control), indicating the presence of biosurfactant. But the drop remained dome shaped for the phosphate buffer which served as the negative control.29
Biosurfactant Mediated Degradation of Crude Oil
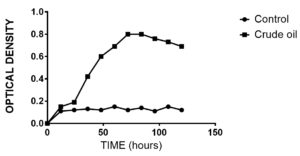
Oil spills are the major routes of contamination of water bodies and coastal environments. Bioremediation is an appealing methodology to remediate these hydrocarbon pollutants. A significant challenge with the remediation of hydrocarbon molecules is its reduced accessibility to the remediating agents. The bigger the exterior space of the oil available to the remediating agents, the quicker the oil slick can be debased. Recent research also confirmed that biosurfactants can decrease the interfacial tension between the molecules and hence the solubility of the hydrocarbon compounds increases.39,40 In the present work, growth of the organism was monitored at regular intervals over 8 days. The highest growth was reached after 72 hrs (Figure 10). Crude oil was supplemented as the sole carbon source. The maximum bacterial growth corresponded to highest biosurfactant production and crude oil biodegradation.41
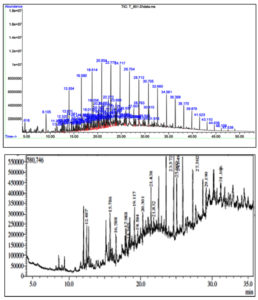
Figure 11. Chromatogram showing the peaks of the crude oil before (top)and after biodegradation (bottom)
From the GC-MS data various hydrocarbons ranges from C1 to C70 are present in the sample(Figure 11). Hydrocarbons analyzed were 3,5-Di-t-butylphenol (C11H13O), Eicosane (C20H42), Hexadecane (C16H34), 2-methyloctacosane (C29H60), Nonahexacontanoic acid (C69H138O2 , 2,6,10,15-tetramethyl Heptadecane (C21H44), Tritetracontane (C43H88), Eicosane10-methyl (C21H44), Sulfurous acid butyl heptadecyl ester (C21H44O3S), Pentadecane 2-methyl (C16H34), Nonadecane 2-methyl (C20H42) , Diisooctyl phthalate (C24H38O4), Eicosane 2-methyl (C21H44), 1-chloro Heptacosane (C27H55Cl). The data was compared with reported crude oil composition. Bolade et al.,42 reported the presence of wide range of aliphatic and aromatic compounds in the crude petroleum sample. This includes BTEX (29%), Acyclic hydrocarbons (22%), PAH (18%), Cyclic hydrocarbons (17%) and others 14%. None of the crude oil components were detected in our analysis in its pure form. In the biodegradation process, high molecular weight compounds get reduced to its low molecular intermediates. As the incubation time increases, some of the petroleum hydrocarbons converted into either intermediate or end products which are considered as secondary metabolites.42 Secondary metabolites like Hexane, 2,3-dimethyl, Heptane, 3-methyl, Cyclohexane, 1,1-dimethyl, Octane, 2-methyl etc. having importance in medicine, dyes and also have antimicrobial, anti-fungal properties.43
A chrysene degrading bacterium, Bacillus halotolerans was screened for its potential to produce biosurfactants. The production medium for biosurfactant production was optimized by suitable modification in the MS medium through RSM approach. The BS production was improved at a pH of 8, nitrogen concentration of 1250 mg/mL, carbon concentration of 75 mg/mL and incubation after 3 days. From the result of characterization, we could conclude that the biosurfactant belongs to the lipopeptide group. The biosurfactant produced was effective in the degradation of crude oil hydrocarbons. In the biodegradation process some of the petroleum hydrocarbons are converted into either intermediate or end products which are considered as secondary metabolites. Secondary metabolites like Hexane, 2,3- dimethyl, Heptane, 3-methyl, Cyclohexane, 1,1-dimethyl, Octane, 2-methyl etc. having importance in medicine, dyes and also have antimicrobial, anti-fungal properties.
ACKNOWLEDGMENTS
The authors would like to thank Department of Biotechnology, Sahrdaya College of Engineering and Technology and Department of Biotechnology, Karunya University, Coimbatore, India for their kind support and encouragement.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
ST and KS conceptualized and designed the study. GJ and ST performed the experiments. ST performed interpretation of results and drafted the manuscript. All authors read and approved the final manuscript for publication.
FUNDING
None.
DATA AVAILABILITY
The datasets generated and/or analysed during the current study are available at Genbank Accession number MK439524.
ETHICS STATEMENT
This article does not contain any studies on human participants or animals performed by any of the authors.
- Sarachat T, Pornsunthorntawee O, Chavadej S, Rujiravanit R. Purification and concentration of a rhamnolipid biosurfactant produced by Pseudomonas aeruginosa SP4 using foam fractionation. Bioresour Technol. 2010;101(1):324-330.
Crossref - Le Guenic S, Chaveriat L, Lequart V, Joly N, Martin P. Renewable Surfactants for Biochemical Applications and Nanotechnology. J Surfactants Deterg. 2019;22(1):5-21.
Crossref - Cooper DG, Goldenberg BG. Surface-active agents from two Bacillus species. Appl Environ Microbiol. 1987;53(2):224-229.
Crossref - Desai JD, Banat IM. Microbial production of surfactants and their commercial potential. Microbiol Mol Biol Rev. 1997;61(1):47-64.
Crossref - Banat IM. Potentials for use of biosurfactants in oil spills cleanup and oil bioremediation. Water Stud. 2000;8:177-185.
- Cooper DG, Macdonald CR, Duff SJ, Kosaric N. Enhanced production of surfactin from Bacillus subtilis by continuous product removal and metal cation additions. Appl Environ Microbiol. 1981;42(3):408-412.
Crossref - Jarvis FG and Johnson MJ, A Glyco-lipide Produced by Pseudomonas Aeruginosa. J Am Chem Soc. 1949;71(12):4124-4126.
Crossref - Burger MM, Glaser L, Burton RM. Formation of rhamnolipids of Pseudomonas aeruginosa. Methods Enzymol. 1966;8:441-445.
Crossref - Guerra-Santos LH, Kappeli O, Fiechter A. Dependence of Pseudomonas aeruginosa continuous culture biosurfactant production on nutritional and environmental factors. Appl Microbiol Biotechnol. 1986;24(6):443-448.
Crossref - Cutler AJ, Light RJ. Regulation of hydroxydocosanoic acid sophoroside production in Candida bogoriensis by the levels of glucose and yeast extract in the growth medium. J Biol Chem. 1979;254(6):1944-1950.
Crossref - Cooper DG, Paddock DA. Production of a biosurfactant from Torulopsis bombicola. Appl Environ Microbiol. 1984;47(1):173-176.
Crossref - Kretschmer A, Bock H, Wagner F. Chemical and physical characterization of interfacial-active lipids from Rhodococcus erythropolis grown on n-alkanes. Appl Environ Microbiol. 1982;44(4):864-870.
Crossref - James K, Kuni T. Effects of Ethambutol. Antimicrobial Agents Chemother. 1981;20(3):401-404.
Crossref - Casas JA, Garcia De Lara S, Garcia-Ochoa F. Optimization of a synthetic medium for Candida bombicola growth using factorial design of experiments. Enzyme Microb Technol. 1997;21(3):221-229.
Crossref - Sarubbo LA, Farias CBB, Campos-Takaki GM. Co-utilization of canola oil and glucose on the production of a surfactant by Candida lipolytica. Curr Microbiol. 2007;54(1):68-73.
Crossref - Thanomsub B, Watcharachaipong T, Chotelersak K, Arunrattiyakorn P, Nitoda T, Kanzaki H. Monoacylglycerols: Glycolipid biosurfactants produced by a thermotolerant yeast, Candida ishiwadae. J Appl Microbiol. 2004;96(3):588-592.
Crossref - Konoshi M, Fukuoka T, Morita T, Imura T, Kitamoto D. Production of new types of sophorolipids by Candida batistae. J Oleo Sci. 2008;57(6):359-369.
Crossref - Alejandro C-S, Hernandez H, Jaramillo-Flores M. Production of glycolipids with antimicrobial activity by Ustilago maydis FBD12 in submerged culture. Afr J Microbiol Res. 2011;5(17):2512-2523.
Crossref - Chandran P, Das N. Biosurfactant Production and Diesel Oil Degradation By Yeast Species Trichosporon Asahii Isolated From Petroleum Hydrocarbon Contaminated Soil. Int J Eng Sci Technol. 2010;2(12):6942-6953.
- Rufino RD, Sarubbo LA, Campos-Takaki GM. Enhancement of stability of biosurfactants produced by Candida lipolytica using industrial residue as substrate. World J Microbiol Biotechnol. 2007;23(5):729-734.
Crossref - Elenga-Wilson PS, Kayath CA, Mokemiabeka NS, Nzaou SAE, Nguimbi E, Ahombo G. Profiling of Indigenous Biosurfactant-Producing Bacillus Isolates in the Bioremediation of Soil Contaminated by Petroleum Products and Olive Oil. Int J Microbiol. 2021;2021:9565930.
Crossref - Salihu A, Abdulkadir I, Almustapha MN. Potential Development on Biosurfactants. Biotechnol Mol Biol Rev Vol. 2009;3(5):111-117.
- Thomas S, Veettil NT, Subbiah K. Isolation, characterization and optimization of chrysene degradation using bacteria isolated from oil-contaminated water. Water Sci Technol. 2021;84(10-11):2737-2748.
Crossref - Masaaki M, Yoshihiko H, Tadayuki I. Masaaki Morikawa, Yoshihiko Hirata, Tadayuki Imanaka. Biochim Biophys Acta. 2000;(1488):211-218.
Crossref - Abouseoud M, Maachi R, Amrane A, Boudergua S, Nabi A. Evaluation of different carbon and nitrogen sources in production of biosurfactants by Pseudomonas fluorescens. Desalination. 2008;223(1-3):143-151.
Crossref - Kaushik R, Saran S, Isar J, Saxena RK. Statistical optimization of medium components and growth conditions by response surface methodology to enhance lipase production by Aspergillus carneus. J Mol Catal B Enzym. 2006;40(3-4):121-126.
Crossref - Chander CRS, Lohitnath T, Kumar DJM, Kalaichelvan PT. Production and characterization of biosurfactant from Bacillus subtilis MTCC441 and its evaluation to use as bioemulsifier for food bio – preservative. 2012;3(3):1827-1831.
- Nitschke M, Pastore GM. Production and properties of a surfactant obtained from Bacillus subtilis grown on cassava wastewater. Bioresour Technol. 2006;97(2):336-341.
Crossref - Venkatesan TV, Murty VR. Production of a Lipopeptide Biosurfactant by a Novel Bacillus sp. and Its Applicability to Enhanced Oil Recovery . ISRN Microbiol. 2013;2013:621519.
Crossref - Qiao N, Shao Z. Isolation and characterization of a novel biosurfactant produced by hydrocarbon-degrading bacterium Alcanivorax dieselolei B-5. J Appl Microbiol. 2010;108(4):1207-1216.
Crossref - Al-Sayegh A, Al-Wahaibi Y, Joshi S, Al-Bahry S, Elshafie A, Al-Bemani A. Bioremediation of Heavy Crude Oil Contamination. Open Biotechnol J. 2016;10(1):301-311.
Crossref - Thaniyavam J, Roongasawang N, Kameyama T, et al. Production and characterization of biosurfactants from Bacillus licheniformis F2.2. Biosci Biotechnol Biochem. 2003;(67):1239-1244.
Crossref - Phulpoto IA, Yu Z, Hu B, et al. Production and characterization of surfactin-like biosurfactant produced by novel strain Bacillus nealsonii S2MT and its potential for oil contaminated soil remediation. Microb Cell Fact. 2020;19(1):1-12.
Crossref - Joy S, Rahman PKSM, Sharma S. Biosurfactant production and concomitant hydrocarbon degradation potentials of bacteria isolated from extreme and hydrocarbon contaminated environments. Chem Eng J. 2017;317:232-241.
Crossref - Meena KR, Sharma A, Kumar R, Kanwar SS. Two factor at a time approach by response surface methodology to aggrandize the Bacillus subtilis KLP2015 surfactin lipopeptide to use as antifungal agent. J King Saud Univ – Sci. 2020;32(1):337-348.
Crossref - Janek T, Gudina EJ, Polomska X, et al. Sustainable surfactin production by Bacillus subtilis using crude glycerol from different wastes. Molecules. 2021;26(12).
Crossref - De Faria AF, Teodoro-Martinez DS, De Oliveira Barbosa GN, et al. Production and structural characterization of surfactin (C14/Leu7) produced by Bacillus subtilis isolate LSFM-05 grown on raw glycerol from the biodiesel industry. Process Biochem. 2011;46(10):1951-1957.
Crossref - Parthipan P, Preetham E, Machuca LL, Rahman PKSM, Murugan K, Rajasekar A. Biosurfactant and degradative enzymes mediated crude oil degradation by bacterium Bacillus subtilis A1. Front Microbiol. 2017;8:1-14.
Crossref - Ankulkar RS, Chavan M. Studies on rhamnolipid biosurfactants by Leclercia Adecarboxylata in crude oil biodegradation. Int J Recent Sci Res. 2020;11(01):36950-36954
- Araujo SC da S, Silva-Portela RCB, de Lima DC, et al. MBSP1: a biosurfactant protein derived from a metagenomic library with activity in oil degradation. Sci Rep. 2020;10(1):1-13.
Crossref - Sharma S, Pandey LM. Production of biosurfactant by Bacillus subtilis RSL-2 isolated from sludge and biosurfactant mediated degradation of oil. Bioresour Technol. 2020;307:123261.
Crossref - Bolade OP, Adeniyi KO, Williams AB, Benson NU. Remediation and optimization of petroleum hydrocarbons degradation in contaminated water using alkaline activated persulfate. J Environ Chem Eng. 2021;9(4).
Crossref - Primeia S, Inoue C, Chien MF. Potential of biosurfactants’ production on degrading heavy oil by bacterial consortia obtained from Tsunami-induced oil-spilled beach areas in Miyagi, Japan. J Mar Sci Eng. 2020;8(8):577.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.