ISSN: 0973-7510
E-ISSN: 2581-690X
Cryptococcus is a non-motile, gram positive, non-fermenting Basidiomycetous encapsulated yeast like fungus that causes respiratory, neurological and other systemic diseases in both humans and animals. Present study delineates the possible distribution of Cryptococcus species in pigeon droppings, excreta of other avian species, eucalyptus tree and contaminated soil specimens collected from different geographical co-ordinates of six geographical regions of the lower Brahmaputra Valley of Assam, India. The fungi were isolated through conventional methods of Sabouraud Dextrose Agar (SDA) and Bird Seed Agar (BSA) media and identified through negative staining of capsule as well as performing classical bio-chemical tests. Identity of the isolates was further confirmed through sequence analysis of ITS-1 and ITS-4 region of the 18S rDNA. Two pathogenic species of Cryptococcus were isolated from 67 (15.40%) of the 435 specimens. Of these positive isolates 41 (9.43%) belonged to Naganishia albida (Cryptococcus albidus) while 26 (5.98%) represented Papiliotrema laurentii (Cryptococcus laurentii). Both the species were recovered from 58 (18.35%) dry and 9 (7.56%) moist specimens. The percentage of prevalence of Naganishia albida in dry and moist specimens were 35 (11.07%) and 6 (5.04%) respectively. Contrary to this, the percentage of prevalence of Papiliotrema laurentii in dry and moist were 23 (7.28%) and 3 (2.52%) respectively. The findings indicate that Cryptococcus species have established a better ecological sustenance in dry specimens than moist. The findings of the investigation demonstrated that the prevalence of Cryptococcus albidus in attics, dovecotes / houses of pigeon fanciers, contaminated soil, eucalyptus tree and droppings of other birds were 11(12.36%) out of 89, 23(14.11%) of 163,2(3.23%) of 62,4(7.84%) of 51 and only 1(1.43%) out of 7 specimens respectively. The recovery of Papiliotrema laurentii in the above specimens were 3(3.37%), 20(12.27%), 1(1.61%), 1(1.96%) and 1(1.42%) respectively. The findings revealed that the prevalence of Naganishia albida is more than that of Papiliotrema laurentii in natural substrates. The notorious pathogenic fungi, Cryptococcus neoformans could not be isolated, indicative of the fact that the region selected for the study is not environmentally favorable for growth and sustenance of the species. Findings of the study clearly demonstrate the ecological and epidemiological significance of the non-neoformans species of the genus cryptococcus that needs further comprehensive studies to access the prevalence of the genus from public health point of view.
Cryptococcus, pigeon droppings, natural substrates, ecological relationship, lower Brahmaputra valley, prevalence
Cryptococcus is non-motile, gram positive, non-fermenting Basidiomycota from the order Tremellale under the family of Tremellaceae of capsulated yeast like fungus 1. Littman (1959) reported the first case of cryptococcosis in man directly attributed to pigeon excreta 2. Outbreaks of cryptococcosis are reported in human, animals and avian species whilst presence of the causative organism has been reported in reptiles, fruits and vegetables 3. Of more than 70 species of the genus Cryptococcus, a majority thrive in the environment and only few of them are medically important disease causing pathogenic agents 4. Amongst all the species, Cryptococcus neoformans is regarded as the major human and animal pathogen, while Cryptococcus albidus and Cryptococcus laurentii are occasionally known to cause moderate to severe diseases especially in immuno-compromised patients 5.
Cryptococcus species are commonly found in dropping of pigeons that apparently harbors the organism in normal commensal form 6. In addition, the digestive tracts of parrots and canaries also harbour the fungus 7. Pigeon droppings, are available in most unlikely places such as roofs and ventilations of abandoned buildings, cornices, leaves and branches of trees that serve as ecological niche for adaptation, dispersion of Cryptococcus replication and transmission. Other environmental sources such as fruits and vegetables retain the fungi as saprophyte that may cause infection in man and animals either by inhalation of spores of the organism or through subcutaneous inoculation 3,8–10.
Excreta of pigeon as saprobic reservoir of Cryptococcus species have been frequently recovered from various countries of the world 3,11,12. Isolation of Cryptococcus from many tree species in America 13 Brazil 14, India 15, Iran 16 have been reported.
Among the systemic and opportunistic mycoses, cryptococcosis continues to exacerbate severe health risks, especially in high risk groups and immuno-compromised patients. It has been estimated that the global prevalence of cryptococcosis among AIDS patients stands at 2.33% and 6.8% both worldwide and within India respectively 3. However, incidence of cryptococcosis in recent years is alarmingly increasing at a global scale amounting to one million infections and approximately 6,25,000 deaths annually, rendering the disease to be considered as an most important fungal diseases 17.
Cryptococcosis is a highly infectious and enigmatic mycotic disease that affect a variety of animals too 18,19. The disease occurs in acute, sub-acute or in chronic forms and have global significance 20. The fungi causes respiratory as well as neurological problems and is often sporadic in nature 19,21,22. Incidentally, the exact epidemiological data on the incidence and prevalence of the disease is not readily available as the disease is not a notifiable one although several reports of considerable morbidity and mortality in human as well as in animals are well documented 21.
The first report of Cryptococcosis in India was documented in 1952 and subsequently many other investigators reported its occurrence from time to time in various parts of the country 23. Although numerous studies on the zoonotic importance and epidemiology of Cryptococcus neoformans have been reported, yet there is lack of proper information as literatures are scanty on the non-neoformans species like Cryptococcus albidus, Cryptococcus laurentii or Cryptococcus uniguttulatus 7. In recent years, a burst of increase in opportunistic infection by these non-neoformans pathogenic yeasts have been observed 24. Environmental sources including canopy leftovers from some trees are regarded as the main sources of Cryptococcus albidus, Cryptococcus laurentii, and Cryptococcus uniguttulatus that lead to infection in human and animals 16,25,26.
Epidemiology of non-neoformans Cryptococcus albidus, Cryptococcus laurentii, Cryptococcus uniguttulatus, Cryptococcus luteolus and some other species of the genus are highly relevant as these species often turn out to be pathogenic and thereby increases the risk of infection. A case of encephalitis in an HIV patient due to Cryptococcus albidus was reported in China 27 while fungal keratitis due to the same species was reported by Huang et.al, (2015) 28. Earlier, pulmonary Cryptococcosis due to Cryptococcus laurentii in diabetis and patients suffering from ganglio-neuroblastoma were reported by Averbuch et.al, (2002) 29 and Shankar et.al, (2006) 30 respectively. Reports are also available on Cryptococcal meningitis, Cryptococcal myelitis and multiple skin lesions in HIV patients infected with Cryptococcus neoformans var. grubii from Assam having a history of occupational exposure to pigeons and chicken 31.
Such findings are of greater significance to places like Assam where domestication of pigeons, fowls and various birds of the finch family are common among the inhabitants at a large scale both as household pets and for meat. A proper surveillance of the sites of occurrence of the fungi as well as characterization of its diverse forms for creating a database is a primary need since the practice of rearing pigeons is non-scientific, traditional and sporadic reports of pulmonary diseases (including cryptococcal meningitis) have been recorded from time to time amongst the common mass. There is a remarkable gap of information on this aspect in the entire northeastern part of India, including Assam. This is a preliminary attempt for identifying the hot spots of Cyptococcus occurrence in environmental samples representing the lower Brahmaputra valley of Assam and characterizing the isolates for their proper identification to generate a baseline data. It is envisaged that the study will generate information for future planned research activities on the opportunistic systemic mycotic disease causing fungi.
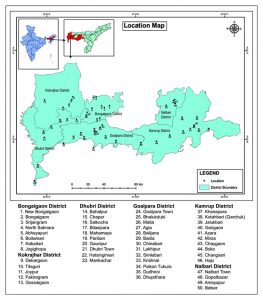
In the present investigation, a total of 435 samples of pigeon droppings and droppings of other avian species, barks and leaf litter of natural stands of eucalyptus trees as well as environmental soil from different sources from five different distinct regions of the lower Brahmaputra Valley of Assam were collected. These included the districts of Bongaigaon, Dhubri –(both north bank extending up to West Bengal border and south bank extending upto the border of the state of Meghalaya along the river Brahmaputra), the district of Kokrajhar BTAD region, the district of Goalpara extending up to the border with Meghalaya, the district of greater Kamrup (including both urban and rural extensions along the north bank of the river Brahmaputra) and finally the district of Nalbari of western lower Assam. Samples collected were transported to the laboratory in ice cooled boxes packed in polyethylene bags. However, cloacal samples of pigeon were collected using sterile cotton swab in a transport media (Eurotubor, Rubi Barcelona, Spain). After collection, all samples were either processed on same day or stored at 40C until being processed for isolating Cryptococcus species. In the meantime, basic environmental characteristics of the collection areas were noted for understanding the ecological and epidemiological relevance of the organism collected from different sources (Table.1, Fig.1, Fig.2 and Fig.3). The marked samples were processed as per standard methods mentioned elsewhere with slight modifications 14,32.
Table (1):
Frequency distribution of samples as per their sources and types.
| Breakup of samples from different sources and geographical regions | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sl. No. | Sample Code | Area/ Region/ District | Geographical coordinates/ Place of Sampling | Season/Month/ Year | Sources of Samples | Total Sample Dry & Moist | ||||||||||||
| Pigeons Droppings | Other Avian Species (Chicken, Ducks etc.) (C) |
Aviary/ Canary cages/ Pet Shop (D) |
Soil contaminated with droppings of Pigeons (E) | Eucalyptus trees (Leaves, Barks and Debris around the trees) (F) |
||||||||||||||
| Attics, /roofs ventilators of old buildings (A) | Pigeon fanciers households (Cloacal/Fresh) (B) | |||||||||||||||||
| Dry | Moist | Dry | Moist | Dry | Moist | Dry | Moist | Dry | Moist | Debris | Barks | Leaves | ||||||
| 1 | BNG | Bongaigaon | Jogighopa & North Salmara, Abhayapuri, Baitamari, Bongaigaon Town, Kabaitari, Chalantapara | July, August, September, October/2018 and March/2017 | 8 | 2 | 18 | 5 | 3 | 3 | 2 | 2 | 7 | 3 | 2 | 2 | 2 | Dry-42 Moist-17 Total = 59 |
| 2 | KJR | Kokrajhar | Jaipur, Kokrajhar Town, Titaguri, Devorgaon | July, August, September/2018 and Feb, March/2017 | 8 | 4 | 15 | 6 | 2 | 2 | 2 | 2 | 4 | 3 | 2 | 2 | 1 | Dry-35 Moist-18 Total = 53 |
| 3 | DHB | Dhubri and South Salmara, Mankachar | Bahalpur, Chapar, Bilasipara, Gauripur, Dhubri Town, Hatsingimari (Meghalaya Border), Mahamaya temple area | June, July, September, October/2018 and Feb, March/2017 | 21 | 3 | 29 | 8 | 3 | 2 | 3 | 3 | 7 | 4 | 3 | 2 | 3 | Dry-68 Moist-23 Total = 91 |
| 4 | KRP | Kamrup Rural and Urban | Mirza, Kotahbari, Gorchuk, Azara, Hajo, Jalukbari, Chaygaon, Boko | June, July, August, September, October/2018 and February/2017 | 11 | 4 | 19 | 4 | 3 | 1 | 3 | 4 | 7 | 4 | 7 | 4 | 4 | Dry-54 Moist-21 Total = 75 |
| 5 | GLP | Goalpara | Lakhipur, Simlabari, Tikrikilla, Meghalaya border, Agia, Krishnai, Dudhnoi,Matia, Dhupdhara and Goalpara | March/2017 and June, July, August, September/2018 | 19 | 3 | 34 | 7 | 4 | 3 | 4 | 5 | 10 | 4 | 6 | 3 | 2 | Dry-80 Moist-24 Total = 104 |
| 6 | NLB | Nalbari | Nalbari town, Gopal Bazar, Belsor, Chamata, Amayapur. | Sep, Oct/2018 and Feb, March, April/2017 | 5 | 1 | 16 | 2 | 3 | 4 | 3 | 4 | 6 | 3 | 2 | 2 | 2 | Dry-37 Moist-16 Total = 53 |
| Total | 72 | 17 | 131 | 32 | 18 | 15 | 17 | 20 | 41 | 21 | 22 | 15 | 14 | Dry-316 Moist-119 Total = 435 |
||||
Fig. 1. Representation of sample collection areas developed from GPS coordinates and using GIS software prepared by Aaranyak, Guwahati, Assam
Isolation and Identification
Processing of samples was done in the Department of Biotechnology, Gauhati University. Two grams (2g) of the samples was diluted in 20 ml of sterile physiological saline and the mixture was stirred for five minutes in a vortex apparatus and allowed to stand for 15-20 minutes for decantation. After this, 1 ml of the supernatant was inoculated onto 9 ml of sterile physiological saline supplemented with chloramphenicol (0.05mg/ml) and then incubated at 370C for an hour. An aliquot of 0.1ml of each supernatant of the processed samples were aspirated and streaked onto duplicate plates of Sabouraud Dextrose Agar (SDA) and a Bird Seed Agar (BSA) supplemented with chloramphenicol (0.05mg/ml).
Cultures with SDA media were incubated at 370C while that of Bird Seed Agar (BSA) were incubated at 250C for 15 days and monitored daily from 2nd day onward for observing growth and presence of colony and to evaluate colony morphology. Brown pigmented colonies were identified as Cryptococcus neoformans while colonies showing cream colouration with smooth and mucoid appearance were identified as non-neoformans species coglomerate. Results evaluated on the basis of morphological data were expressed as the average nos. of viable yeast cells per gram of sample (CFU/g). For comparisons, Cryptococcus laurentii MTCC 2898 (procured from CSIR Institute of Microbial technology, Chandigarh – 1600361, India) was used as control. Identified isolates were sub cultured, purified and further subjected to an array of morphological, cultural and bio-chemical tests such as carbohydrates, nitrite reduction, urease production and phenol oxidation 11,15,16,20,24.
Capsule Identification
For capsule identification of the probable Cryptococccus spp., a drop of inoculum from newly inoculated isolates grown on SDA was added onto India ink stain on a sterile glass slide with a cover slip and observed under a bright field microscope (Olympus CX33) at 100 and 400 times magnification (Kwon-Chung and Bennett, 1992) and also phase contrast microscopy (Leica DM750) under 1000 fold magnification. Presence of distinct wide round to oval gelatinous capsule, with or without hyphae was considered as positive observation 33–36.
Molecular Characterization
DNA extraction
For DNA extraction, isolates from pure cultures were inoculated onto 1.5ml Eppendorf tubes containing 0.5 ml of Sabouraud Dextrose broth supplemented with chloramphenicol and incubated overnight in an orbital shaker at 150 rpm and 30°C. After 24 hours, the fungal suspensions with predetermined concentrations were centrifuged at 5,000 rpm for 10 minutes, and the pellet was frozen at -20°C for 1 hour with further incubation at 65°C for 1 h in 0.5 ml of extraction buffer containing 50 mM Tris-HCl, 50 mM EDTA, 3% sodium dodecyl sulfate and 1% 2-mercaptoethanol (Ferrer et al., 2001). The lysate was finally extracted with phenol-chloroform-isoamyl alcohol (v/v) in the ratio 25:24:1. To this, 65μl of 3M sodium acetate and 75μl of 1M NaCl were added and the resulting volume was incubated at 4°C for 30 minutes. DNA was recovered by isopropanol precipitation and washed with 70% (v/v) ethanol. Concentration of DNA was measured at 260 nm in a UV-VIS Spectrophotometer (Shimadzu) and stored at -20°C until further use 37–39.
PCR amplification
For characterization of Cryptococcus species, isolated DNA was amplified in a gradient PCR (Eppendorf Nexus Gradient). The primers used for amplification included the D1/D2 regions targeting ITS1 and ITS4 with expected fragment length of 600 bp. Details of the primers are given () in Table 2. The program for amplification was set for 1 cycle of initial denaturation for 30 minutes at 940C followed by 35 cycles of denaturation for 1 minute at 940C, primer annealing for 1 minute at 550C, chain extension for 1 minute at 720C and a final extension for 7 minutes at 720C respectively. The PCR products after amplification were resolved in a 1.5 % agarose gel subjected to electrophoresis and was visualized under UV gel documentation system (UVitec Cambridge, Genei). The amplicons were later stored at -200C for further analysis 40–43.
Table (2):
Sequence of primers used for PCR amplification.
Primer |
Sequence |
|---|---|
ITS1 |
3′- TCC GTA GGT GAA CCT GCG G -5′ |
ITS4 |
5′- TCC TCC GTC TAT TGA TAT GC -3′ |
Sequencing
Sequencing of PCR products were done at Xcelris Labs Limited, India through outsourcing. Chromatogram files obtained were analyzed for nucleotide-BLAST on NCBI portal for identification of the species. Phylogram and dendrogram was prepared using PhyML (Phylogenetic Maximum Likelihood) (http://www.atgc-montpellier.fr/phyml/).
Statistical analysis
A very brief descriptive analysis was carried out to interpret the data as they were mostly qualitative attributes. However, positive results obtained from different sources were compared through k-proportion test through Monte Carlo / Marascuilo methods to assess their homogeneity across sources with the help of XLSTAT software 44–46.
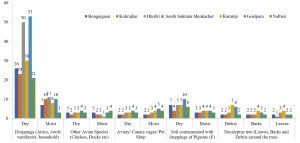
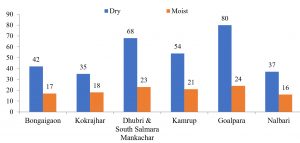
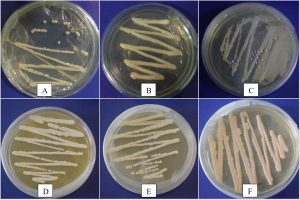
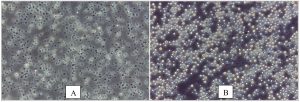
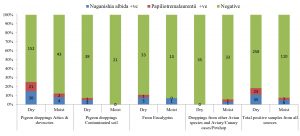
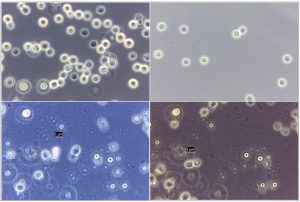
In the present study, a total of 67 Cryptococcus spp were recovered from 435 samples with a prevalence of 15.40% (Table 3). Out of 67 positive isolates, 41 (9.43%) were identified as Cryptococcus albidus (Naganishia albida) Table 3, Fig. 4, Plate-D, E, F while 26 (5.98%) were identified as Cryptococcus laurentii (Papiliotrema laurentii). Details of the identified (Table. 3, Fig. 4, Plate-B, C). Cultural and biochemical characteristics of Naganishia albida and Papiliotrema laurentii were almost similar except in the utilization of potassium nitrate which was negative in case of Papiliotrema laurentii. Cultural characteristics of Cryptococcus laurentii (Fig. 4, Plate B, C) in SDA media were comparable with the reference strain MTCC 2898 (Fig. 4, Plate A). Based on the type of sample (dry and moist), Naganishia albida (Cryptococcus albidus) and Papiliotrema laurentii (Cryptococcus laurentii) could be isolated from 58 (18.35%) of dry and 9 (7.56%) of moist specimens collected from different geographical niches / co-ordinates. (Table 3, Fig. 7).
Table (3):
Frequency distribution of positive samples of Cryptococcus species (pooled sample).
| Species of Cryptococcus | Pigeon droppings with code no. | Pigeon droppings contaminated with soil (C) |
From Eucalyptus Trees (D) |
Droppings from other avian species and aviary/Canary cages/ Pet shops (E) |
Total positive samples from all sources (F) | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (A) | (B) | (A+B) | ||||||||||||||||||||
| Attics/Ventilators/roofs of private and public places / old and abandoned buildings (nos. and %) |
Houses of Pigeon fanciers/dovecotes (nos. and %) | Pigeon droppings Attics & dovecotes (nos. and %) |
Nos. and % | Nos. and % | Nos. and % | Nos. and % | ||||||||||||||||
| Dry | Moist | Total | Dry | Cloacal/ Moist |
Total | Dry | Moist | Total | Dry | Moist | Total | Debris (dry) | Barks (dry) | Leaves (moist) | Total | Dry | Moist | Total | Dry | Moist | Total | |
| Total no. of samples | 72 | 17 | 89 | 131 | 32 | 163 | 203 | 49 | 252 | 41 | 21 | 62 | 22 | 15 | 14 | 51 | 35 | 35 | 70 | 316 | 119 | 435 |
| Naganishia albida (positive & %) | 10 (13.89) | 1 (5.88) | 11 (12.36) | 20 (15.27) | 3 (9.37) | 23 (14.11) | 30 (14.78) | 4 (8.16) | 34 (13.49) | 2 (4.88) | 0 (0.00) | 2 (3.22) | 3 (13.64) | 0 (0.00) | 1 (7.14) | 4 (7.84) | 0 (0.00) | 1 (2.85) | 1 (1.42) | 35 (11.07) | 6 (5.04) | 41 (9.43) |
| Papiliotrema laurentii (positive & %) |
3 (4.17) | 0 (.00) | 3 (3.37) | 18 (13.74) | 2 (6.25) | 20 (12.27) | 21 (10.34) | 2 (8.08) | 23 (9.13) | 1 (2.44) | 0 (0.00) | 1 (1.61) | 0 (0.00) | 1 (6.67) | 0 (0.00) | 1 (1.96) | 0 (0.00) | 1 (2.85) | 1 (1.42) | 23 (7.28) | 3 (2.52) | 26 (5.98) |
| Naganishia albida &Papiliotrema laurentii (positive & %) |
13 (18.05) | 1 (5.88) | 14 (15.73) | 38 (29.07) | 5 (15.62) | 43 (26.38) | 51 (25.12) | 6 (12.24) | 57 (22.61) | 3 (7.32) | 0 (0.00) | 3 (4.84) | 3 (13.64) | 1 (6.67) | 1 (7.14) | 5 (9.80) | 0 (0.00) | 2 (5.71) | 2 (2.85) | 58 (18.35) | 9 (7.56) | 67 (15.40) |
Fig. 4. ((A) Reference strain ) Papiliotrema laurentii (MTCC 2898), (B, C) Papiliotrema laurentii, (D, E, F) Naganishia albida.
Fig. 5. Phase contrast microscopy of reference sample – Papiliotrema laurentii (MTCC 2898), under 100X magnification
Fig. 6. Phase contrast microscopy of (A) Papiliotrema laurentii (B) Naganishia albida under 40x magnification.
Fig. 7. Distribution of positive samples of Cryptococcus species isolated from samples and specimens
During the investigation of 57 (22.61%) Cryptococcus species isolated from pigeon droppings from sites A & B, 34 (13.49%) were identified to be Cryptococcus albidus while 23 (9.13%) represented Cryptococcus laurentii. The highest recovery percentage of Cryptococcus (57) was observed in pigeon droppings and the least (2) was from avian species other than pigeon. In terms of relative isolation of positive cultures of Cryptococcus, 14 (15.73%) were from attics / ventilations / old and abandoned buildings (Site A), 43 (26.38%) were from houses of pigeon fanciers / dovecots (Site B), 3(4.84%) were from contaminated environment / soil (Site C), 5 (9.80%) were from the debris / barks / leaves of eucalyptus trees (Site D) while 2 (2.86%) were from other avian sources like chicken, duck, parrot etc (Site E). The results are presented in Table 3.
Table (4):
Isolates of Cryptococcus Species Naganishia albida (cryptococcus albidus) and Papiliotrema laurentii(cryptococcus laurentii) from different sources of samples.
| Sl. No. | Collection site | Environmental niches | Dry | Moist | Total | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| n | Positive | Relative (absolute) isolations (No. & %) |
n | Positive | Relative & absolute isolations/ proportions (No. & %) |
N | Positive | Relative & absolute isolations/ proportions (No. & %) |
|||
| 1 | Attics / ventilations of old abandoned buildings (A) | Pigeons droppings (Columba livia) | 72 | 13 | 18.05 (4.11) | 17 | 1 | 5.88 (0.84) | 89 | 14 | 15.73 (3.22) |
| 2 | Houses of Pigeon fanciers / dovecotes (B) | Pigeons’ droppings (Columba livia) | 131 | 38 | 29.00 (12.02) | 32 | 5 | 15.62 (4.20) | 163 | 43 | 26.38 (9.89) |
| 3 | Environmental samples/ soil ( C) | Contaminated with pigeon droppings | 41 | 3 | 7.32 (0.95) | 21 | 0 | 0 | 62 | 3 | 4.84 (0.69) |
| 4 | Eucalyptus trees (D) | Debris, barks (dry) and leaves (moist) | 37 | 4 | 10.81 (1.27) | 14 | 1 | 7.14 (0.84) | 51 | 5 | 9.80 (1.15) |
| 5 | Other birds (E) |
Parrots, chicken, ducks etc. | 35 | 0 | 0.00 | 35 | 2 | 5.71 (1.66) | 70 | 2 | 2.86 (0.46) |
| 6 | Total samples (F) | 316 | 58 | 18.35 | 119 | 9 | 7.56 | 435 | 67 | 15.40 | |
Figures in parenthesis indicate percent with respect to total sample size (N), n = number of samples for both dry and moist samples.
* P = 0.001 (<0.05)
Relative and absolute percentage of isolates of Cryptococcus (Naganishia albida and Papiliotrema laurentii) from each collection site was also analyzed. Prevalence rate of isolates from Site A (3.22%), Site B (9.89%), Site C (0.69%), Site D (1.15%) and Site E (0.46%) was recorded (Table 4). Results interpret that there exist a significant difference at 0.05% among the variants of different proportions obtained from various samples during the study. Values having the superscript across different rows do not differ significantly.
Table (5):
Isolates of Naganishia albida (Croptococcus albidus) from different sources of samples.
| Sl. No. | Collection site / Sources | Environmental niches | Collected samples/ Positive isolates | Absolute isolations/ relative isolations (No. & %) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dry | Moist | Total | ||||||||||
| 1 | Attics / ventillations of old / abandoned / building (A) | Pigeons’ droppings (Columba livia) | n | Positive | Relative & absolute isolations/ isolations (No & %) |
n | Positive | Relative & absolute isolations/ isolations (No & %) |
N | Positive | Relative & absolute isolations/ isolations (No & %) |
|
| 72 | 10 | 13.89 / 3.16 | 17 | 1 | 5.88/0.84 | 89 | 11ab | 12.36 / 2.52 | ||||
| 2 | Houses of Pigeons’ fancies / dovecotes (B) | Pigeons’ droppings (Columba livia) | 131 | 20 | 15.27 / 6.33 | 32 | 3 | 9.37/2.52 | 163 | 23b | 14.11 / 5.29 | |
| 3 | Environmental samples/ soil (C) | Contaminated with pigeon droppings |
41 | 2 | 4.88 / 0.63 | 21 | 0 | 0.00 | 62 | 2a | 3.23 / 0.46 | |
| 4 | Eucalyptus tree (D) | Debris and barks (dry) and leaves (moist) |
37 | 3 | 8.10 / 2.59 | 14 | 1 | 7.14/0.84 | 51 | 4ab | 7.84 / 0.22 | |
| 5 | Other birds (E) | Parrots, chicken, ducks etc. |
35 | 0 | 0.00 | 35 | 1 | 2.85/0.22 | 70 | 1a | 1.43 / 0.84 | |
| 6 | Samples Total (F) | 316 | 35 | 11.07 | 119 | 6 | 5.04 | 435 | 41 | 9.43 | ||
Figures in parenthesis indicate percent with respect to total sample size (N), ‘n’ indicate number of samples (Values having the superscript across different rows do not differ significantly)
* P = 0.008 (<0.05)
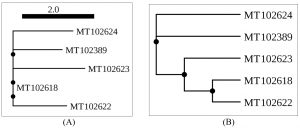
The overall prevalence of Naganishia albida, across all dry and most environmental samples was 9.43% (41 of 435). However, the prevalence of the species was 35 (11.07%) in dried specimens in comparison to 6 (5.04%) from moist specimens. Details of prevalence of Naganishia albida is presented in Table 5 whilst morphological features of the species is presented in the Fig 4 (D, E, F) as well as Fig. 6B, and Fig. 8 respectively. Results interpret a significant difference at 0.05 % level among the different proportions obtained from different sources during the study. On the contrary, the overall prevalence of Papiliotrema laurentii in all dried and moist environmental samples were 5.98% (26 of 435), 7.28% (23 of 316) and 2.52% (3 of 119) respectively from different sites. Details of the prevalence of the species is presented in Table. 6, Fig. 6(A) and Fig. 9 alongwith the comparable datasheet of reference strain (Cryptococcus laurentii MTCC 2898, Fig. 5). Result correlate a significant difference at 0.05 % level among the representative proportions obtained from different sources during the study. Details of genetic identity of the isolated strains of Naganishia albida and Papiliotrema laurentii including sequenced data of the rDNA ITS region and their accession numbers obtained from NCBI after submission of the sequences are presented in Table. 7 and Fig. 10 respectively.
Table (6):
Isolates of Papiliotrema laurentii (Cryptococcus laurentii) from different sources of samples.
| Sl. No | Collection site | Environmental niches | Collected samples/ Positive isolates |
Absolute isolations/ relative isolations (No. & %) |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Dry | Moist | Total | |||||||||
| n | Positive | Relative & absolute isolations/ isolations (No. & %) |
n | Positive | Relative & absolute isolations/ isolations (No. & %) |
N
|
Positive | Relative & absolute isolations/ isolations (No. & %) |
|||
| 1 | Attics / ventillations of old / abandoned / building (A) | Pigeons’ droppings (Columba livia) |
72 | 3 | 4.17/0.95 | 17 | 0 | 0.00 | 89 | 3 | 3.37/0.69 |
| 2 | Houses of Pigeons’ fancies / dovecotes (B) | Pigeons’ droppings (Columba livia) |
131 | 18 | 13.74 / 5.70 | 32 | 2 | 6.25 (0.69) | 163 | 20 | 12.27 / 4.60 |
| 3 | Environmental samples/ soil ( C) | Contaminated with pigeons’ droppings |
41 | 1 | 2.44 / 0.32 | 21 | 0 | 0.00 | 62 | 1 | 1.61 / 0.23 |
| 4 | Eucalyptus tree (D) | Debris and barks (dry) and leaves (moist) |
37 | 1 | 2.70/0.32 | 14 | 0 | 0.00 | 51 | 1 | 1.96 / 0.23 |
| 5 | Other birds (E) |
Parrots, chicken, ducks etc. |
35 | 0 | 0.00 | 35 | 1 | 2.85 (0.84) | 70 | 1 | 1.42 / 0.23 |
| 6 | Samples Total (F) | 316 | 23 | 7.28 | 119 | 3 | 2.52 | 435 | 26 | 5.98 | |
Figures in parenthesis indicate percent with respect to total sample size (N), ‘n’ indicate number of sample (Values having the superscript across different rows do not differ significant) * P = 0.001 (<0.05)
Table (7):
Sequence data of Cryptococcus species isolated from different sources.
Sl No |
Laboratory Code |
Source |
Spacies identified (isolates) |
Accession No |
Sequence (in FASTA format) |
|---|---|---|---|---|---|
1 |
GLP 19 |
Pigeon (Columba livia) dropping |
Naganishia albida (Cryptococcus albidus) |
MT 102389 |
>MT102389.1 Naganishia albida isolate GLP19 small subunit ribosomal RNA gene, partial sequenceTTCTGGTGCCCAAGCAGCCGCGGTAATTCCAGCTCCAATTAGCGTATATT AAAGTTGTTGCAGTTAAAAAGCTCGTAGTTGAACTTCAGGCCCGACGGGGTGGTCTGCCTC ACGGTATGTACTATCCGGTTGGGCCTTACCTCCTGGTGAGCCCGTATGTCGTTTACTCGGTG TGCGGGGGAACCAGGAATTTTACTTTGAAAAAATTAGAGTGTTCAAAGCAGGCATATGCCCGA ATACATTAGCATGGAATAATAGAATAGGACGTGCGGTTCTATTTTGTTGGTTTCTAGGATCGCC GTAATGATTAATAGGGACGGTTGGGGGCATTAGTATTCAGTTGCTAGAGGTGAAATTCTTAGA TTTACTGAAGACTAACTACTGCGAAAGCATTTGCCAAGGACGTTTTCATTAATCAAGAACGAA GGTTAGGGGATCAAAAATGATTAGATACCGTTGTAGTCCTAACAGTAAACTATGCCGACTAG GGATCGGGCCATGTTCAACTTTTGACTGGCTCGGCACCTTACGAGAAATCAAAGTCTTTGGG TTCTGGGGGGAGTATGGTCGCAAGGCTGAAACTTAAAGGAATTGACGGAAGGGCACCACC AGGCGTGGAGCCTGCGGCTTAATTTGACTCAACACGGGGAAACTCACCAGGTCCAGACATA GTAAGGATTGACAGATTGATAGCTCTTTCTTGATTCTATGGGTGGTGGTGCATGGCCGTTCTT AGTTGGTGGAGTGATTTGTCTGGTTTAATTCCGATAACGAAACGAGACCTTTAACCTTGCGT TAGAATTAGGACCCCGGGTTCAGGGGACCT |
2 |
DHB 39 |
Evironmental sample near pigeons’dovecotes (contaminated soil) |
Papiliotrema laurentii (Cryptococcus laurentii) |
MT 102618 |
>MT102618.1 Papiliotrema laurentii isolate DHB39 small subunit ribosomal RNA gene, partial sequenceTTTCTGGTGCCGCAGCAGCCGCGGTAATTCCAGCTCCAGTAGCGTATAT TAAAGTTGCTTGCAGTTAAAAAGCTAGTAGTCGAACTTCGGCCCTGGCTGGACGGTCCGCC TTACGGTGTGCACTGTCCGGCCGGGTCTTACCCTCTTGGTGAGGCCGCATGCCCTTTACTGG GTGTGCGGTGGAACCAGGAATTTTACCTTGAGAAAATTAGAGTGTTCAAAGCAGGCATTTGCC CGAATACATTAGCACGGAATAATAGAATAGGACGTGCGGTTCTATTTTGTTGGTTTCTAGGATC GCCGTAATGATTAATAGGGACGGTCGGGGGCATTAGTATTCCGTTGCTAGAGGTGAAATTTTTA GATTTACGGAAGACTAACTTCTGCGAAAGCATTTGCCAAGGACGTTTTCATTGATCAAGAACG AAGGTTAGGGGATCAAAAACGATTAGATACCGTTGTAGTCTTAACAGTAAACTATGCCGACTAG GGATCGGGCCACGTTAATTTCTGACTGGCTCGGCACCTTACGAGAAATCAAAGTCTTTGGGT TCTGGGGGGAGTATGGTCGCAAGGCTGAAACTTAAAGGAATTGACGGAAGGGCACCACC AGGTGTGGAGCCTGCGGCTTAATTTGACTCAACACGGGGAAACTCACCAGGTCCAGACATA GTAAGGATTGACAGATTGATAGCTCTTTCTTGATTCTATGGGTGGTGGTGCATGGCCGTTCTT AGTTGGTGGAGTGATTTGTCTGGTTAATTCCGATAACGAACGAGACCTTAACCTGCTAAATAG CCAGGCCGGCTTTGGCTGGTTCCGGTACTTCTTTTACTGGACTGTACGTCTATTATACCCACA CTGGATCTGGGACTAACCGCCGGTCGCTAGATCCCCCCTATGAGA |
3 |
KRP 21 |
Dry barks of eucalyptus tree (Eucalyptus camaldulensis) |
Papiliotrema laurentii (Cryptococcus laurentii) |
MT 102622 |
>MT102622.1 Papiliotrema laurentii isolate KRP21 small subunit ribosomal RNA gene, partial sequenceTTCCCGCTGCTGCTGCCCGCCCCGCACGCGGCAATCCAGCTCCAGTAGCGTAT ATTAAAGTTGTTGCAGTTAAAAAGCTCGTAGTCGAACTTCGGGCCTGGCTGGACGGTCCGCCTT ACGGTGTGCACTGTCCGGCCGGGTCTTACCTCTTGGTGAGGCCGCATGCCCTTTACTGGGTGT GCGGTGGAACCAGGAATTTTACCTTGAGAAAATTAGAGTGTTCAAAGCAGGCATTTGCCCGAAT ACATTAGCATGGAATAATAGAATAGGACGTGCGGTTCTATTTTGTTGGTTTCTAGGATCGCCGTA ATGATTAATAGGGACGGTCGGGGGCATTAGTATTCCGTTGCTAGAGGTGAAATTCTTAGATTTAC GGAAGACTAACTTCTGCGAAAGCATTTGCCAAGGACGTTTTCATTGATCAAGAACGAAGGTTAGG GGATCAAAAACGATTAGATACCGTTGTAGTCTTAACAGTAAACTATGCCGACTAGGGATCGGGCC ACGTTAATTTCTGACTGGCTCGGCACCTTACGAGAAATCAAAGTCTTTGGGTTCTGGGGGGAGT ATGGTCGCAAGGCTGAAACTTAAAGGAATTGACGGAAGGGCACCACCAGGTGTGGAGCCTGCG GCTTAATTTGACTCAACACGGGGAAACTCACCAGGTCCAGACATAGTAAGGATTGACAGATTGA TAGCTCTTTCTTGATTCTATGGGTGGTGGTGCATGGCCGTTCTTAGTTGGTGGAGTGATTTGTC TGGTTAATTCCGATAACGAACGAGACCTTAACCTGCTAAATAGCCAGGCCGGCTTTGGCTGGT CGTCGGCTTCTTAGAGGGACTGTCGGCGTTTAGCCGACGGAGTTTGAGCAATCACAGATATAA |
4 |
BNG 4 |
Pigeon (Columba livia) dropping |
Papiliotrema laurentii (Cryptococcus laurentii) |
MT 102623 |
>MT102623.1 Papiliotrema laurentii isolate BNG4 small subunit ribosomal RNA gene, partial sequenceGCGGTAATTCCAGCTCCAGTAGCGTATATTAAAGTTGTTGCAGTTAAAAGCTCG TAGTCGAACTTCGGGCCTGGCTGGACGGTCCGCCTTACGGTGTGCACTGTCCGGCCGGGTCTT ACCTCTTGGTGAGGCCGCATGCCCTTTACTGGGTGTGCGGTGGAACCAGGAATTTTACCTTGA GAAAATTAGAGTGTTCAAAGCAGGCATTTGCCCGAATACATTAGCATGGAATAATAGAATAGGA CGTGCGGTTCTATTTTGTTGGTTTCTAGGATCGCCGTAATGATTAATAGGGACGGTCGGGGGC ATTAGTATTCCGTTGCTAGAGGTGAAATTCTTAGATTTACGGAAGACTAACTTCTGCGAAAGCATT TGCCAAGGACGTTTTCATTGATCAAGAACGAAGGTTAGGGGATCAAAAACGATTAGATACCGTT GTAGTCTTAACAGTAAACTATGCCGACTAGGGATCGGGCCACGTTAATTTCTGACTGGCTCGGC ACCTTACGAGAAATCAAAGTCTTTGGGTTCTGGGGGGAGTATGGTCGCAAGGCTGAAACTTAA AGGAATTGACGGAAGGGCACCACCAGGTGTGGAGCCTGCGGCTTAATTTGACTCAACACGGG GAAACTCACCAGGTCCAGACATAGTAAGGATTGACAGATTGATAGCTCTTTCTTGATTCTATGGG TGGTGGTGCATGGCCGTTCTTAGTTGGTGGAGTGATTTGTCTGGTTAATTCCGATAACGAACGA GACCTTAACCTGCTAAATAGCCAGGCCGGCTTTGGCTGGTCGTCGGCTTCTAGAGGGACGTCG GCGTTTAGCCGACGGAAGTTTGAGGCAATAACA |
5 |
KJR 27 |
Pigeon (Columba livia) dropping |
Naganishia albida (Cryptococcus albidus) |
MT 102624 |
>MT102624.1 Naganishia albida isolate KJR27 small subunit ribosomal RNA gene, partial sequenceAGCTCCAATAGCGTATATTAAAGTTGTTGCAGTTAAAAAAGCTCGTAGTTGAAC TTCAGGCCCGACGGGGTGGTCTGCCTCACGGTATGTACTATCCGGTTGGGCCTTACCTCCTG GTGAGCCCGTATGTCGTTTACTCGGTGTGCGGGGGAACCAGGAATTTTACTTTGAAAAAATT AGAGTGTTCAAAGCAGGCATATGCCCGAATACATTAGCATGGAATAATAGAATAGGACGTGC GGTTCTATTTTGTTGGTTTCTAGGATCGCCGTAATGATTAATAGGGACGGTTGGGGGCATTA GTATTCAGTTGCTAGAGGTGAAATTCTTAGATTTACTGAAGACTAACTACTGCGAAAGCATTT GCCAAGGACGTTTTCATTAATCAAGAACGAAGGTTAGGGGATCAAAAATGATTAGATACCGTT GTAGTCCTAACAGTAAACTATGCCGACTAGGGATCGGGCCATGTTCAACTTTTGACTGGCTCG GCACCTTACAAAAAATCAAATTCTTTGGGTTCTGGGGGAAGTATGGTCGCAAGGCTGAACCT TAAAAGAATTGACGGAGGGGCACCACCAGGCTTGAAGCCGGCGGCTTATTTTAACTCATCACG GGGAAACTCACCAGGTCCGACACATACTAGGATTGACACATTGATATTTCTTTCTTGATTCTC TGGGTGGCGGTGCATGCCCGTTCTAAGTTGGTGGAGTGATTTGTCTGGTTAATGCCCAATCA CGAACGTACATCTTAACCTGCTAACTACACCGATCGGGCTTTGAGCTGCACCCTCTATTCTTTA |
Table (8):
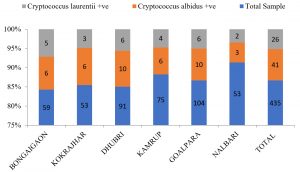
Geographical co-ordinates and environmental niches wise frequency distribution of positive isolates of Cryptococcus species.
| Sl No. | Geographical region | Total Sample | Naganishia albida positive (nos.) |
Papiliotrema laurentii positive (nos.) |
Total positive sample (nos.) | Relative % in the region | Absolute % in the region |
|---|---|---|---|---|---|---|---|
| 1 | BONGAIGAON | 59 (BNG 1-59) | 6 | 5 | 11 | 18.64 | 2.53 |
| 2 | KOKRAJHAR | 53 (KJR 1-53) | 6 | 3 | 9 | 16.98 | 2.06 |
| 3 | DHUBRI | 91 (DHB 1-91) | 10 | 6 | 16 | 17.58 | 3.68 |
| 4 | KAMRUP | 75 (KRP 1-75) | 6 | 4 | 10 | 13.34 | 2.99 |
| 5 | GOALPARA | 104 (GLP 1-104) | 10 | 6 | 16 | 15.38 | 3.68 |
| 6 | NALBARI | 53 (NLB 1-53) | 3 | 2 | 5 | 9.43 | 1.15 |
| P = 0.762 (>0.05) : NS | |||||||
| TOTAL = 435 | Total = 41 | Total = 26 | 67 | 15.04 % | |||
NS: indicate non-significant
Comparisons pertaining to the prevalence of Naganishia albida and Papiliotrema laurentii across the studied geographical locations depict an overall 11 (18.64%), 16 (17.58%), 9 (16.98%), 16 (15.38%). 10 (13.33%) and 5 (9.43%) numbers of positive isolates representing the districts of Bongaigaon, Dhubri, Kokrajhar, Goalpara, Kamrup and Nalbari (Table. 8; Fig. 11). We could not establish any significant difference in the results observed across different districts. Moreover, the prevalence of pathogenic Cryptococcus neoformans could not be observed from any of the tested samples indicating absence of the species in the specimens collected which may be due to unfavorable habitat niche for the species along the lower Brahmaputra valley of Assam resulting in complete absence of the most feared representative form of the family of Cryptococcus.
Cryptococcosis is a major public health concern in India. Epidemiology and pathogenicity of this enigmatic myco-zoonosis in man and animals have been well studied 18–21. Isolation and identification of Cryptococcus from the environment demonstrates the importance of bio-diversity and environmental niche of the pathogenic species and strains of this basidiomycota yeast type fungus because such species usually increase the risk of infection in susceptible population exposed to hypersensitive reactions. Besides, vulnerability of infection among the immuno-compromised hosts other than the infectious state of different biovars of this pathogen, includes both pathogenicity and antifungal resistance which have tremendous implications. Ecological relationship of Cryptococcus was not known until Emmons (1955)10 found the fungus in droppings of pigeon and soil colonized by the genera. Later, his observation was substantiated to be true by many researchers 2,3,17,20,24.
Although many studies have been conducted on the epidemiology and pathogenicity of Cryptococcus neoformans, yet there is scarcity of current information on the prevalence, epidemiology and pathogenicity of the non-neoforman species. The present study defines the incidence of non-neoforman species of the genus Cryptococcus inherent in pigeon droppings, ecological niche of eucalyptus trees and in other microfloral sources of the environment at a specific geographical region along the lower Brahmaputra valley of Assam.
Our observation confirmed the presence of two non-neoforman species of the genus Cryptococcus, viz., Naganishia albida and Papiliotrema laurentii. To our knowledge, this is the first report on the isolation of these two species from pigeon droppings and other environmental sources of the northeastern region of India and this may be considered as an established fact that the environmental sources of the studied areas serve as ideal saprobic reservoirs of these two opportunistic pathogens. Considering the importance of the species at the backdrop of health status of man and animals, these observations may be considered important as the prevalence of cryptococcosis is currently at a rising trend and an increase in the cases of fungemia both in human and animals have been on the rise in the past few years. However, in the current investigation, not a single isolate of Cryptococcus neoformans was recovered despite our incessant attempts to culture the isolate in appropriate Bird Seed Agar and Sabouraud Dextrose Agar media as per international standards.
The overall prevalence of Cryptococcus spp. was 15.40% (67) retrieved from all six geographical regions studied. Of the 67 positive isolates, 9.43% (41) and 5.98% (26) were identified to represent Naganishia albida and Papiliotrema laurentii respectively. The results are comparable with other findings reported by Jang et al (2011)26 who stated the occurrence of 14.30% Cryptococcus albidus and 7.9% Cryptococcus laurentii in pigeon droppings from China. In another study, Kamari et.al, (2017)16 reported 33 of 186 (17.7%) positive cultures and 11 out of 88 (12.5%) confirmed cultures of Cryptococcus spp. retrieved from pigeon nests and eucalyptus tree specimens from Ilam province in Iran. The authors also documented the prevalence of Cryptococcus albidus (17.2%), C. albidus var. kuetzingii (3.4%), C. adilensis (3.4%), C. uzbekistanensis (3.4%) and C. neoformans var grubii (3%) from pigeon nests while they concluded the presence of only C. adilensis (25%) in specimens from eucalyptus trees. .
The finding of 23 (14.11%) Cryptococcus in pigeon droppings from the dovecotes / houses of pigeon fanciers was comparable with the study of Kamari et. al, (2017)16. Our findings revealed 21 (10.34%) positive isolates of Papiliotrema laurentii in the droppings of pigeon in dovecotes /fanciers whereby 4 out of 51 (7.84%) and 1 (1.96%) represented Naganishia albida and Papiliotrema laurentii respectively. Similarly, 20.60% positive isolation rate of Cryptococcus in 6 out of 186 pigeon droppings in attics was reported earlier by Kamariet.al, (2017). In this study, 38 out of 89 (15.73%) isolates from attics were positive for Cryptococcus spp. The slight variation of the findings might be due to differences of climate, humidity, temperature and other biotic or abiotic factors characteristic of both the countries that are geographically apart and separated by a huge landmass.
In the current study, 13.49% (34) and 9.13% (23) of Naganishia albida and Papiliotrema laurentii could be recovered from pigeon droppings from attic ventilations of old buildings, dovecotes houses of pigeon fanciers and from cloacal swabs. Earlier, Rosario et. al. (2009)24 reported a lower isolation rate of 23 (6.9%) Cryptococcus albidus and 2 (0.6%) Cryptococcus laurentii from droppings of pigeons in Spain which may be due to differences in environmental and climatic conditions as mentioned above. Studies have also been reported from Nigeria whereby Pal (2015) 47, had isolated 16 representatives of Cryptococcus neoformans from environmental samples of pigeon droppings with a prevalence of 12.5%. In the present study, the prevalence of Naganishia albida (13.49%) and Papiliotrema laurentii (9.13%) in pigeon droppings and in eucalyptus tree specimens were comparable to previous reported findings, despite the absence of Cryptococcus neoformans.
Although Cryptococcus neoformans is regarded as the most commonly occuring species of the Cryptococcus family, its absence in probable specimens collected from this part of the country is quite intriguing and it reflects the status of overall persistence of the species under humid conditions exposed to abrupt change in temperatures, a feature commonly observed in the studied locations. . The effect of seasonal variations on the persistence of non-neoforman Cryptococcus isolates cannot be nullified and it suggest the need of proper screening through a robust molecular approach concomitant to the behavior and food sources of the birds questioned, not to mention the presumable role of temperature and other edaphic factors that needs further appreciation.
Isolation of several species and varieties of the genus Cryptococcus from different sources, soils, atmospheric airs, dust and even from other avian species i.e chicken, ducks, parrots and munia had been previously reported by many authors. In a study conducted in Brazil, Leite et. al, (2012)48 found the occurrence of 18 (21.4%) Cryptococcus species from 84 specimens of dust collected from public libraries. The isolation rate of Cryptococcus albidus 2 (4.6%) in their study was found to match our findings from dry soil 2 (4.88%). Furthermore, the percentage of isolation of Cryptococcus species in this study was 18.33% (58) as compared to 7.56% (9) from a total of 435 collected specimens in dry and moist conditions. The observations were in congruence with the number of isolates from pigeon droppings whereby 25.12% (51) and 12.24% (6) Cryptococcus isolates could be recovered from dry and moist specimens.
While studying the frequency of isolation in context of regional characteristics, no specific trend in terms of frequency was observed as the number of isolates varied from 9.43% (in Nalbari region) to 18.64% (Bongaigaon region), despite close proximity of the study areas that share the same environmental conditions. It may be recalled that factors like humidity, rainfall, temperature and pH of a definite geographical location do affect population parameters of domesticated birds like pigeons which need further evaluation. Ecological relevance of the non-neoformans species of Cryptococcus (4.84% in soil, 9.80% in eucalyptus tree samples) was the most important findings of this study with profound influence on replication and transmission of the pathogenic species from the ideal sources to the susceptible population at risk. Such situation is very imminent and highly dangerous for crowded and highly populated areas like Nalbari and Kamrup as the route of entry of such microorganisms is either through inhalation or direct skin contact.
From this investigation, it may be concluded that Naganishia albida and Papiliotrema laurentii are the most prevalent Cryptococcus species in the lower Assam belt compared to the more pathogenic Cryptococcus neoformans indicating the need of extensive epidemiological studies for establishing proper identify of the different strains that perpetuate, thrive and adapt to changing environmental conditions which are bound to challenge public health and pose as potential hazards in the near future. It may be stated that to ensure safety of the people, mass awareness programmes on the prevalence of the fungus need to be arranged whilst proper prevention, control and management of such disease causing pathogens be prioritized to prevent future outbreaks so as to minimize the risk of transmission of opportunistic Cryptococcus infections.
ACKNOWLEDGMENTS
The authors duly acknowledge the support and contribution provided by Department of Biotechnology, Gauhati University, Guwahati, Assam to carry out the research activity. The authors also duly acknowledge the support and cooperation received from state department of Animal husbandry and Veterinary, Govt of Assam, Xcelris Labs Limited and other personnel involved during the data collection / processing to carry out task effectively.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
ETHICS STATEMENT
Not applicable. However, during the sample collection from pigeons, proper handling was done as per the standard veterinary practices.
AVAILABILITY OF DATA
As per the research design, data were collected from different primary sources / generated are available with the researcher and NCBI database (Accession No: MT 102389, MT 102618, MT 102622, MT 102623, MT 102624)
- Lewis JL, Rabinovich S. The wide spectrum of cryptococcal infections. Am J Med. 1972;53(3):315-322.
Crossref - Littman ML. Cryptococcosis (Torulosis). Current concepts and therapy. Am J Med. 1959;27(6):976-998.
Crossref - Pal M, Dave P. Cryptococcosis: An Emerging Airborne Mycosis of Global Concern. Air & Water Borne Diseases. 2016;5(1):1000127.
Crossref - Kwon-Chung KJ, Boekhout T, Wickes BL, Fell JW. Systematics of the Genus Cryptococcus and Its Type Species C. neoformans. Cryptococcus. 2014.
- Cheng MF, Chiou CC, Liu YC, Wang HZ, Hsieh KS. Cryptococcus laurentii fungemia in a premature neonate. J Clin Microbiol. 2001;39(4):1608-1611.
Crossref - Darze C, Lucena R, Gomes I, Melo, A. Clinical and laboratory characteristics of 104 Cryptococcus meningoencephalitis cases. Revista Da Sociedade Brasileira de Medicina Tropical. 2000.
- Rosario I, Acosta B, Colom F. La paloma y otras aves como reservorio de Cryptococcus spp Pigeons and other birds as a reservoir for Cryptococcus spp. Revista Iberoamericana de Micologia. 2008;25(1):S13-S18.
Crossref - Emmons CW. Isolation of Cryptococcus neoformans from soil. J Bacteriol. 1951;62(6):685-690.
Crossref - Emmons CW. Prevalence of Cryptococcus neoformans in pigeon habitats. Public Health Rep. 1960:75:362-364.
Crossref - Emmons CW. Saprophytic sources of Cryptococcus neoformans associated with the pigeon (Columba livia). Am J Epidemiol. 1955;62(3):227-232.
Crossref - Ruiz A, Velez D, Fromtling RA. Isolation of saprophytic Cryptococcus neoformans from Puerto Rico: Distribution and variety. Mycopathologia. 1989;106(3):167-170.
Crossref - Fromtling RA, Abruzzo GK, Ruiz A. Virulence and antifungal susceptibility of environmental and clinical isolates of Cryptococcus neoformans from Puerto Rico. Mycopathologia. 1989;106(3):163-166.
Crossref - Lazera MS, Cavalcanti MAS, Londero AT, Trilles L, Nishikawa MM, Wanke B. Possible primary ecological niche of Cryptococcus neoformans. Med Mycol. 2000;38(5):379-383.
Crossref - Filiu WFdeO, WankeIII B, Aguena SM, et al. Cativeiro de aves como fonte de Cryptococcus neoformans na cidade de Campo Grande, Mato Grosso do Sul, Brasil. Revista da Sociedade Brasileira de Medicina Tropical. 2002;35(6).
Crossref - Kumar CPG, Prabu D, Mitani H, Mikami Y, Menon T. Environmental isolation of Cryptococcus neoformans and Cryptococcus gattii from living trees in Guindy National Park, Chennai, South India: Letter to the editor. Mycoses. 2010;53(3):262-264.
Crossref - Kamari A, Sepahvand A, Mohammadi R. Isolation and molecular characterization of Cryptococcus species isolated from pigeon nests and Eucalyptus trees. J Mycol Med. 2017;3(2):20-25.
Crossref - Pal M. Cryptococcus gattii: an emerging global mycotic pathogen of humans and animals. J Mycopathol Res. 2014.
- Pal M, Randhawa HS. Caprine mastitis due to cryptococcus neoformans. Med Mycol. 1976;14(3):261-263.
Crossref - Pal M, Mehrotra BS. Ciyptococcal Mastitis in Daily Animals/Cryptococcen-Mastitis bei Milchvieh. Mycoses. 1983;26(12):615-616.
Crossref - Pal M. First report of isolation of Cryptococcus neoformans var. neoformans from avian excreta in Kathmandu, Nepal. Revista Iberoamericana de Micologia. 1997.
- Mitchell TG, Perfect JR. Cryptococcosis in the Era of AIDS-100 Years after the Discovery of Cryptococcus neoformans. Clin Microbiol Rev. 1995;8(4):515-548.
Crossref - Casadevall A, Mukherjee J, Scharff MD. Monoclonal antibody based ELISAs for cryptococcal polysaccharide. J Immunol Methods. 1992;154(1):27-35.
Crossref - Chander J, Sapra RK, Talwar P. Incidence of cryptococcosis in and around Chandigarh, India, during the period 1982-91. Mycoses. 1994;37:23-26.
Crossref - Rosario I, de Mendoza MH, Deniz S, Soro G, Alamo I, Acosta B. Isolation of Cryptococcus species including C. neoformans from cloaca of pigeons. Mycoses. 2005:48(6):421-424.
Crossref - Bernardo FM, Martins HM, Martins L. Fontes urbanas de Cryptococcus spp – Lisboa Urban Sources of Cryptococcus spp – Lisbon ( Portugal ). Revista Portuguesa de Ciencias Veterinarias. 2001.
- Jang YH, Lee SJ, Lee JH, Chae HS, Kim SH, Choe NH. Prevalence of yeast-like fungi and evaluation of several virulence factors from feral pigeons in Seoul, Korea. Lett Appl Microbiol. 2011;52(4):367- 371.
Crossref - Liu Y, Ma S, Wang X, Xu W, Tang J. Cryptococcus albidus encephalitis in newly diagnosed HIV-patient and literature review. Med Mycol Case Rep. 2014;3:8-10.
Crossref - Huang YH, Lin IH, Chang TC, Tseng SH. Early diagnosis and successful treatment of cryptococcus albidus keratitis. Medicine (United States). 2015;94(19):e885.
Crossref - Averbuch D, Boekhout T, Falk R, et al. Fungemia in a cancer patient caused by fluconazole-resistant Cryptococcus laurentii. Med Mycol. 2002;40(5):479-484.
Crossref - Shankar EM, et al. Pneumonia and pleural effusion due to Cryptococcus laurentii in a clinically proven case of AIDS. Can Respir J. 2006;13(5):275-278.
Crossref - von Geldern G, Nath A. Central nervous system infections. in Evidence-Based Neurology: Management of Neurological Disorders: Second Edition. 2015.
Crossref - Criseo G, Bolignano MS, de Leo F, Staib F. Evidence of canary droppings as an important reservoir of Cryptococcus neoformans. Zentralblatt fur Bakteriologie. 1995;282(3):244-254.
Crossref - Liu Z, Ma L, Zhong Y, Wang X, Xie S. Isolation, identification and significance of Cryptococcus neoformans and Candida albicans from faecal specimen of pigeon. Advances in Intelligent and Soft Computing. 2012;134:507-512.
Crossref - Grossgebauer K. Detection of cryptococcus neoformans capsule using fluorescent, mucopolysaccharide-binding dyes. Klin Wochenschr. 1980;58(18):943-945.
Crossref - Chae HS, Park GN, Kim SH, et al. Rapid direct identification of Cryptococcus neoformans from pigeon droppings by nested PCR using CNLAC1 gene. Poultry Science. 2012;91(8):1983-1989.
Crossref - Filion T, Kidd S, Aguirre K. Isolation of Cryptococcus laurentii from Canada Goose guano in rural upstate New York. Mycopathologia. 2006;162(5):363-368.
Crossref - Mseddi F, Jarboui M, Sellami A, Sellami H, Ayadi A. A rapid and easy method for the DNA extraction from Cryptococcus neoformans. Biol Proced Online. 2011;13(1):5.
Crossref - Tendulkar SR, Gupta A, Chattoo BB. A simple protocol for isolation of fungal DNA. Biotechnol Lett. 2003;25(22):1941-1944.
Crossref - Motkova P, Vytrasova J. Comparison of methods for isolating fungal DNA. Czech Journal of Food Sciences. 2011;29:s76-s85.
Crossref - Leal AL, Faganello J, Bassanesi MC, Vainstein MH. Cryptococcus species identification by multiplex PCR. Med Mycol. 2008;46(4):377-383.
Crossref - Lau A, Chen S, Sorrell T, et al. Development and clinical application of a panfungal PCR assay to detect and identify Fungal DNA in tissue specimens. J Clin Microbiol. 2007;45(2):380-385.
Crossref - Mitchell TG, Freedman EZ, White TJ, Taylor JW. Unique oligonucleotide primers in PCR for identification of Cryptococcus neoformans. J Clin Microbiol. 1994;32(1):253-255.
Crossref - Sidrim JJC, Costa ANF, Cordeiro RA, et al. Molecular methods for the diagnosis and characterization of cryptococcus: A review. Can J Microbiol. 2010;56(6):445-458.
Crossref - XLSTAT. Statistical Power for Repeated Measures Anova. Adinsoft. 2015.
- Barbosa AM, Santos BFdeO, Carvalho EdEO, et al. Biological activity of Cryptococcus neoformans and Cryptococcus gattii from clinical and environmental isolates. Jornal Brasileiro de Patologia e Medicina Laboratorial. 2013;49(3).
Crossref - Adinsoft S. XLSTAT-software, version 10. Addinsoft, Paris, France. 2010.
- Pal M. First Record of Isolation of Cryptococcus neoformans from Pigeon Droppings in Djibouti. Molecular Microbiology Research. 2015;5(3):1-3.
Crossref - Leite DP, Amadio JVRS, Martins ER, et al. Cryptococcus spp isolated from dust microhabitat in Brazilian libraries. Journal of Occupational Medicine and Toxicology. 2012;7(1)11.
Crossref
© The Author(s) 2020. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.