Antibiotics are an essential part of modern healthcare, revolutionizing medicine and saving countless lives worldwide. However, the emergence of antimicrobial resistance (AMR) is a growing concern, with the potential to cause a public health crisis in the future. The aim of this review article is to provide an overview of the microbial and anthropogenic factors contributing to AMR, as well as the consequences of inaction to address the AMR crisis. We searched various international databases such as PubMed, Scopus, ScienceDirect and Google Scholar using “Antimicrobial Resistance”,” Superbug”, “Antibiotic Stewardship”, “One Health’ and “Surveillance” as search keywords in different combinations. We have thoroughly discussed the causes of AMR, such as the overuse and misuse of antibiotics, and the development of resistant strains of bacteria. We have also suggested possible interventions to combat AMR, such as the one health approach, antibiotic stewardship protocols, and the application of artificial intelligence in drug design. Additionally, we have explored the benefits of traditional ethnic medicinal practices in therapy. In conclusion, this review article emphasized the urgent need for a comprehensive and strategic plan to address the issue of AMR. Further in-depth research and novel approaches can mitigate the growing menace of AMR and safeguard both human and animal populations.
Antimicrobial Resistance, Health Care, Antibiotics, Superbug, Surveillance
Antibiotics have been hailed as one of the most remarkable discoveries of the 20th century, credited with saving millions of lives from deadly infectious diseases and nosocomial infections since their serendipitous discovery by Sir Alexander Fleming in the 1920s. However, their effectiveness is being increasingly overshadowed by a phenomenon known as antimicrobial resistance (AMR), which makes frequent use of these drugs an ineffective one. As the great English taxonomist John Ray once said, “Nothing is invented and perfected at the same time”, and unfortunately, this rings true for our promising drugs whose efficacy is being compromised by AMR. With overuse, inaccurate diagnosis, and multiple unnecessary antimicrobial prescriptions, many infectious diseases are now becoming untreatable due to acquired resistance.
AMR is a phenomenon whereby microorganisms such as bacteria, viruses, fungi, and some parasitic organisms gain resistance against the antibiotics used to kill these microorganisms. It is a growing concern that many antibiotics have now become significantly resistant, posing a significant threat to treat the infectious diseases which need urgent attention.1
The review article includes a comprehensive study on the development of antimicrobial resistance, the consequences, and the novel approaches to address the AMR. For this purpose, we searched various international databases such as PubMed, Scopus, ScienceDirect and Google Scholar using “Antimicrobial Resistance”,” Superbug”, “Antibiotic Stewardship”, “One Health’ and “Surveillance” as search keywords in different combinations. Out of the various published articles, 71 articles were discussed with relevance to the present topic. The review article will help the researchers in this field to design their research work with updated information in the field of antimicrobial resistance.
Consequences of antimicrobial resistance
Economic consequences
Antimicrobial resistance (AMR) is a growing concern for countries worldwide, with potentially catastrophic effects on both the economy and healthcare systems. The development of new antibiotics is an expensive and time-consuming process, making them largely inaccessible to lower-income groups.2 In the United States, the outsourcing of resistant infections by immigrants has been identified as a major driver of healthcare expenses, causing a substantial burden on the budget. This trend is partly due to the search for better healthcare and economic opportunities, leading to immigration from countries with poor health infrastructure. Economic simulation studies that were carried by the World Bank predicts that by the end of year 2050, the AMR related loss on the annual GDP will be around 1.1%.3 The silver lining however is that 50% of this projected loss can be avoided by adopting robust and appropriate containment measures. Direct costs of the AMR on the economy will be in form of increased health expenses that will put pressure on the government to increase its health budget overlooking other sectors of economy.
The economic repercussions of AMR are far-reaching, with losses to the workforce and increased morbidity and mortality rates. In addition, some countries have implemented travel restrictions and immigration curbs to prevent the spread of superbug infections. The economic loss in India, due to the AMR has not been documented thoroughly, but the impacts on Indian economy can be huge in the future.4 A recent study on global burden of bacterial AMR, revealed that an estimated 1.27 million deaths were attributable to bacterial AMR.5 A staggering estimate suggests that by 2050, death rates could reach up to 10 million people.6 Furthermore, the impact on international trade of livestock-based products, particularly animal protein sources like milk and meat, will be catastrophic due to the supply-demand mismatch created by the fear of zoonotic diseases spreading from resistant strains of superbugs.7
Impact on healthcare system
The AMR has tremendous impact on human health and more vulnerable groups are the neonates (with increased mortality rates), older adults with weakened immune systems, and immunocompromised individuals from economically disadvantaged backgrounds who cannot afford second-line antimicrobial treatments when the initial treatment fails to cure their resistant infections.
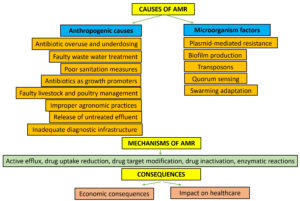
Causes of antimicrobial resistance
AMR is a major challenge at present, faced by the mankind and it spreads through both vertical (one generation to next generation) and horizontal gene transfer (e.g. Transformation, transduction, and conjugation).8 There are various reasons responsible for the development of the AMR (Figure). The different mechanisms are active efflux, reduction of drug uptake, drug target modification, drug inactivation, and by many enzymatic reactions.8,9
The various causes of AMR include plasmid mediated resistance, biofilm development, anthropogenic causes as major factors among many other reasons.
Plasmid-mediated resistance
Development of plasmid mediated resistance accounts to the highest incidences of AMR. Plasmids are that extrachromosomal genetic materials that help in transfer of information from one bacterial cell to another and help in bacterial conjugation sometimes. Plasmids which can acquire genes of antimicrobial resistance and can thereby aid in bacteria becoming resistant. The resistance against colistin has been observed in many Enterobacteriaceae species and the plasmid-mediated mcr-1 gene is found to confer the resistance as reviewed by Rhouma et al.10 With the impact of policy on one health programme the colistin withdrawal as livestock growth promoter, further reduced the existence of colistin-resistant strains and their mcr-1 gene in livestock, human and the environment. The sustainable policy like one health approach is essential to curb the menace of antibiotic resistance in future. The plasmid-mediated gene dissemination through horizontal gene transfer facilitates the distribution of antibiotic resistance genes more prominently as compared to other ways.11 The environmental pollutants such as metals (along with its nanoparticles), pharmaceuticals (antibiotics and non-antibiotics), and microplastics influence the plasmid transfer in aquatic as well as terrestrial systems. Future studies should focus on rapid identification of these pollutants and their effect on the enhancement of plasmid transfer as well as blocking of gene transfer, so that strategies can be made to control antibiotic resistance. Yassine et al. reviewed the prevalence of plasmid-mediated quinolone resistance (PMQR) in the Arab countries and found the increased incidence of quinolone resistance by Enterobacteriaceae bacteria.12 The plasmid-encoded extended-spectrum β-lactamases (ESBLs) and carbapenemases promote antibiotic resistance significantly, as reviewed by Schultsz and Geerlings.13 Some drugs like colistin, Fosfomycin, tigecycline, and temocillin were found effective against carbapenem-resistant strains of Enterobacteria. The plasmid-mediated dissemination of antimicrobial resistance in Salmonella enterica strains has been reviewed by Carattoli.14 It is observed that enteric pathogens gain resistance against expanded-spectrum cephalosporins. The integrons (integration of resistant genes in specialized genetic elements) play crucial role in gaining and disseminating the resistance genes.14 The in vivo study on plasmid-mediated antimicrobial resistance (AMR) by plasmid pOXA-48, conferring resistance to the carbapenems in enterobacterial colonies were observed from the gut samples taken from hospitalized patients.15 The development of quinolone resistance is reviewed by Guan et al. and it is observed that plasmid-mediated resistance, gene mutation, alterations in drug efflux, and alterations in cell membrane activity are the prominent reasons of development of quinolone-resistant bacteria.16 Study on the plasmid-mediated antibiotic resistance among uropathogens of primigravid women with bacteriuria, revealed that 65% of E. coli isolates and 41% of K. pneumoniae isolates to be ESBL positive.17 The genes responsible for the resistance were TEM-1 (66.7%) and CTX-M-15 (33.3%). The study concluded that there is more prevalence of plasmid-mediated ESBL and quinolone resistance microorganisms in community-acquired urinary tract infections of primigravid women.17 Study by Kivata et al. on Neisseria gonorrhoeae isolates from Kenya, revealed the existence of plasmid-mediated penicillin and tetracycline resistance in N. gonorrhoeae and the responsible resistant genes were TEM (penicillin-resistant), and TetM (tetracyclin-resistant) genes.18 The transporters resistance nodulation division (RND) are encoded from chromosomal genes and confer multi-drug resistance in Gram-negative bacteria. Study by Lv et al. on Klebsiella pneumoniae isolates from animal origin, identified a novel RND efflux pump gene cluster (tmexCD1-toprJ1) on plasmids. The antibiotics tigecycline is used generally to treat the carbapenem-resistant K. pneumoniae infection. However, the plasmid-derived RND efflux pump gene cluster (tmexCD1-toprJ1) was found to show resistance against tigecycline which is a major concern.19 The One health approach measures can be adopted to curb the less distributed plasmid-derived tmexCD1-toprJ1 gene cluster in human and animal population. The Gram-negative bacteria Bhargavaea beijingensis is mainly observed in soil and water. The PS04 strain of B. beijingensis is found having anti-microbial resistance through the plasmid repUS12 which carries the resistant genes ermT and tet(L) that provide resistance against macrolides, lincosamides, and tetracycline.20 This study revealed the emergence of the plasmid repUS12 in B. beijingensis bacteria might be through horizontal gene transfer and attention must be given for surveillance of environmental bacteria with relevance to antimicrobial resistance development. Schwarz et al. have discussed thoroughly about the plasmid-mediated resistance in Staphylococcus sp. and Firmicutes.21 Apart from plasmids there are transposons (the mobile genetic elements), commonly referred to as jumping genes. They can change their position within the same DNA and can sometimes confer insertion and transfer of AMR genes which ultimately converts them to an antibiotic resistant superbug.22 The other non-inheritable mechanisms involve swarming adaptations, bacteria growing themselves on biofilms, quorum sensing and persisted cells that are important contributors to the antimicrobial resistant phenotypes.
Role of biofilms on antimicrobial resistance
Biofilms are adaptation strategies for microbial survival. The association of biofilm formation and activation of AMR genes need further exploration. However; many studies revealed that biofilm can function as a barrier in providing resistance against the antibiotics. A comprehensive review done by Datta et al. in the context of Indian patients, showed that biofilms are predominantly formed in bacteria such as Escherichia sp., Streptococcus sp., Staphylococcus sp., and Pseudomonas sp. in patients with respiratory, urinary, dental, and skin infections.23 There is increased multi-drug resistance in those microbes which needs further study. The Salmonella typhi (MTCC-733) biofilm significantly interrupted the antibiotic response and it was observed that the cellulose component of biofilm could be involved in resistance development and can be used as a drug target against the biofilm of S. typhi.24 Study on Mycobacteroides abscessus colony morphology and their impact on biofilm development and antibiotics resistance, revealed that the smooth colony morphotypes (with biofilm) have more antimicrobial resistance, as compared to that of rough colony morphotypes.25 The biofilm mode of growth of these bacteria enhanced the resistance against the antibiotics amikacin and tigecycline. Moreover, these bacteria became more tolerant to the disinfectants peracetic acid. The small molecules and nanoparticles are found to be effective in inhibiting the biofilm production and may work well against microbial resistance, as reviewed by Gattu et al.26 Study by Pauze-Foixet et al. showed that polymyxin B affects biofilm formation in Vibrio cholerae more effectively under anaerobic condition.27 Genetically manipulated bacteriophages, phage proteins, and enzymes can have better effects against superbugs and many antibiotic-resistant bacteria.28 These substances can be used alone or in combination with the antibiotics to combat the antibiotic-resistant bacteria in future.
In Biofilm growth the bacteria rely on the microbial colony and the matrix that reduces the antimicrobial potency by techniques such as diminishing its diffusion within the bacterial biomass and the sometimes the biofilm matrix seizes active principles of antibiotics through its complex structure.29 It is also noteworthy that different regions in biofilms have different physicochemical microenvironments for example oxygen rich areas of Pseudomonas aeruginosa are defenseless against the quinolones whereas contrasting trends seen in non-oxygen rich regions.22 Biofilm limits the chemotherapeutic action of antimicrobials as they prevent the penetration of antibiotics thereby acting as an antibiotic resistant medium.30 Quorum sensing on the other hand is a phenomenon by which bacterial cells regulate their expression of genes in response to fluctuations in cell population density.31 The chemical signals called auto inducers helps the bacteria in an array of roles like motility, sporulation, in swarming behavior and even sometimes in biofilm production to evade antibiotics; although most often quorum sensing acts as a commensal interaction between hosts and pathogenic bacteria. These variety of techniques have motivated the scientists to study the loopholes within most of these mechanisms extensively and thereby developing the strategies to tackle the menace of superbugs.
Anthropogenic causes of antimicrobial resistance
Antibiotic overuse is rampant in both animal and human health sectors. In animals, sub-therapeutic doses of antibiotics are given as growth promoters, leading to a rapid growth rate, and contributing to the development of antibiotic-resistant strains. Similarly, inappropriate use of antibiotics in humans, including under-dosing or incomplete courses, allows bacteria to mutate and develop resistance, posing a grave threat to public health in the form of emerging superbugs. Anthropogenic activities have great impact on the distribution of antibiotic resistance in both clinical environments as well as livestock management, agronomic practices, and waste water treatment.32 The anthropogenic factor such as use of antibiotics in livestock and poultry farming, and wastewater effluents from animal slaughter houses are the major reasons of wide distribution of antimicrobial resistance in the environment. Ciprofloxacin was found to have highest resistance in livestock slaughter houses in Iran (93%), Nigeria (50%), and China (20%), whereas in Germany (21-81%) and Spain (56%) highest resistance of ciprofloxacin observed in poultry slaughter houses.33 Profiling of the AMR (antimicrobial resistance) and virulence factors of Salmonella spp. and Escherichia coli isolated from various anthropogenic areas, showed the resistance of Salmonella spp. to sulfisoxazole and streptomycin; and resistance of E. coli to sulfisoxazole, cefoxitin, and ampicillin.34 The study by Navarro et al. on the antibiotic resistance of bacterial isolates from water samples from a pre-Alpine and an Apennine River (Variola and Tiber River) (low anthropogenic impact area), revealed that blaTEM conferred resistance to β-lactam antibiotics and present in around 58% of isolates as compared to 9% for mefA/E which conferred resistance to macrolides.35 These findings concluded that there is existence of co-resistant bacteria even in the nonanthropogenic environment which might spread to humans and animals in due course of time. Study on the antimicrobial resistance of nontuberculous Mycobacteria from the nosocomial environment, revealed that the M. obuense and M. mucogenicum have multi-drug resistance property.36 The erm (erythromycin resistance methylase) gene confers resistance for M. obuense against clarithromycin; whereas, Corynebacterium isolates were resistant to penicillin, ciprofloxacin and linezolid compounds. Study by Swift et al. on development of AMR in bacterial isolates from wildlife birds and animals, revealed that the AMR is driven by anthropogenic origin, due to exposure of antimicrobials and antimicrobial resistant bacteria.37
Poor sanitation measures facilitate the spread of infectious diseases. The lack of compliance to proper sanitation and hygiene methods is one of the reasons of enhancing the cases of AMR all over the world.38 Apart from these, poor diagnostic infrastructure facilities might fail to accurately diagnose the disease and its underlying causative agent, which in turn paves way for the doctors prescribing the wrong antibiotics that give rise to cross-resistance. Besides, anthropogenic activities such as the use of antibiotics as growth promoters in livestock and poultry, faulty wastewater treatment, and the release of untreated effluent have contributed significantly to the selection pressure driving AMR, triggered by antimicrobial pollutants in waterbodies. Thus, there is a growing concern regarding the role of human activities in fueling the rise of antimicrobial resistance, which has become a critical global public health challenge. Several studies have pointed towards the fact that prevalence of antibiotic resistance genes in effluent treatment plants are always more than the concentration of the same in natural rivers and faulty treatment leads to the dissemination of antibiotic resistance genes in environment.39 The lack of awareness and access to other alternatives to antibiotics at reasonable cost price also contributes to this crisis.
Novel approaches to address AMR and way forward
Multiple synergistic approaches are required to mitigate the Antimicrobial Resistance (AMR) in the current scenario which needs to be properly addressed among all the stake holders through proper awareness. The human gut is a reservoir of many antibiotic-resistant microorganisms and antibiotic therapy can disturb the gut microbiome by changing their taxonomic and functional composition, and promoting pathogen colonization.40,41 The dysbiosis in gut microbiome can lead to increased burden of antibiotic resistant genes, and invasion of antibiotic-resistant pathogens into the blood stream, causing many diseases.42 The functional metagenomics and long-read sequencing technologies can play crucial role in detecting and understanding the antibiotic resistant genes and preparing strategies for treatment against antibiotic-resistant bacteria. New innovative approaches are listed below (Table 1) with their advantages to address AMR. Besides, an overview of techniques are listed in Table 2, which might mitigate the AMR issue.
Table (1):
Novel approaches with their possible advantages to address AMR
Strategy |
Techniques |
Advantages |
|---|---|---|
Newer antibiotic development |
Explore unexplored bacteria for novel antibiotic molecules |
Develop antibiotics that reduce reliance on existing ones |
Anti-virulence strategies |
Use chelators, secretion system inhibitors, sanitizers |
Protect antibiotics, find alternatives to minimize use |
Improved low-cost diagnostics |
Portable devices, centralized sero-surveillance |
Accurate diagnosis, cost-effective healthcare |
Stricter control in agriculture |
Regulations, promote organic farming |
Reduce zoonotic antibiotic-resistant infections |
Antibiotic cycling |
Temporarily withhold and reintroduce susceptible antibiotics |
Preserve effectiveness of susceptible antibiotics |
Pulsed Electromagnetic Field |
Use pulsed EMF with antimicrobial peptides |
Increase antibiotic effectiveness |
One Health Approach |
Integrate human, animal, and environmental health |
Prevent emerging zoonotic superbugs |
Antibiotic Stewardship |
Evidence-based prescribing, incentives for development |
Proper antibiotic use, reduced impact on resources |
Nano antibiotics |
Coat nanoparticles on antibiotics, use adjuvant molecules |
Increase antibiotic efficacy, prevent resistance |
Host body immunomodulation |
Vaccination, probiotics for microbial flora alteration |
Reduce disease severity, alternative to antibiotics |
Traditional medicinal practices |
Use medicinal herbs, promote organic products |
Cost-effective alternative to antimicrobials |
Artificial Intelligence |
Algorithm-based drug design, machine learning |
Targeted drug design, predict emerging superbugs |
Table (2):
Overview of techniques to mitigate the AMR issue
Newer Strategy and Techniques |
Concept and Techniques |
Advantages |
|---|---|---|
1) Newer antibiotics development and discovery |
• Exploring newer antibiotic molecules from unexplored and hard to grow those bacteria in labs by culturing them in vivo by isolation chip mechanism that mimics their natural habitat. |
• Development of newer noble antibiotic compounds that ease the pressure on the existing group of antibiotics |
2) Anti-virulence strategies and modifying the structure of Antimicrobials |
• Use of chelators and secretion system inhibitors to protect antibiotics • Use of alcohol-based sanitisers and disinfectants |
• Protects the antibiotics from antagonistic proteins of bacteria. • Alternatives to antibiotics in elimination of bacteria; minimising need for antimicrobials |
3) Improved and accurate diagnosis, development of low-cost diagnostics |
• Development of low cost precise and portable diagnostic devices along with centralised sero surveillance and data analysis |
• Encourage correct prescribing of antimicrobials, with cost effective accurate disease diagnosis for public |
4) Policy of stricter control over excessive misuse of antibiotics in agriculture and animal husbandry |
• Formulation of strict laws regulations to prevent misuse of antibiotics as growth promoter in poultry and livestock farming. • Promotion of organic farming and antibiotic free animal products |
• Will limit the emergence of zoonotic food borne antibiotic resistant infections |
5) Antibiotics Cycling |
• Not prescribing a particular susceptible class of antibiotics for a limited period of time and then again reintroducing it in future |
Protects a susceptible class of antibiotics from being ineffective due to emergence of antibiotic resistance |
6) Use of Pulsed Electromagnetic Field against bacteria |
• Use of pulsed EMF to sensitize a bacterial cell along with antimicrobial peptide |
• Increases penetration of antimicrobial peptide towards a gram-negative bacterial cell thereby increasing the efficacy of antibiotics and killing it by electroporation |
7) One Health Approach |
• Integration of human health, animal health and environment by promotion of research in emerging zoonotic infections and resistant microbes arising due to climate change |
• Promotion of transdisciplinary research and collection of data and its analysis to prevent emerging zoonotic superbugs |
8) Antibiotic Stewardship |
• Evidence based prescribing of antibiotics and rolling out incentives for development of newer and noble antibiotics, eliminating bottlenecks involved |
• Preventing overuse as well as wrong use of antibiotics by judicious prescribing of antibiotics. • Benefits the patient and minimises impact on the limited antibiotics at dispense. |
9) Nano antibiotics and Antibiotic adjuvant |
• Coating nanoparticles over antibiotics to increase its efficacy. • Use of adjuvant molecules |
Increases the efficacy of antibiotics manifold and effective in preventing emergence of resistance |
10) Host-body immunomodulation |
• Effective and regular vaccination to prevent infection transmission of diseases, reducing severity of diseases • Use of probiotics as a medium to alter microbial flora |
• Monoclonal antibodies as an effective alternative to antibiotics. • Vaccines prevent severe infection and minimise need of antibiotics. • Faecal microbial transplants and probiotics to prevent drug resistant infections. |
11) Traditional and ethnic medicinal and veterinary practices |
• Use of medicinal herbs to treat some infectious diseases • Promotion of organic and antibiotic residue free products |
• Effective as well as relatively inexpensive way to reduce dependence on antimicrobials |
12) Application of Artificial Intelligence and Computational biology along with machine learning |
• Use of algorithm in drug designing • Use of machine learning to predict newer emerging resistant superbugs |
• Whole genome sequencing to design the newer drugs to target pathogens • AI based prediction models to predict superbugs • Use of AI to design drug combinations via 3D printing and antimicrobial peptide modelling |
Newer antibiotics development and discovery
Exploration of antibiotics modules from microbes that live in extreme conditions and have not been exposed yet can be a newer area of hope to tackle AMR. Furthermore, with the advancement of science and nanobiotechnology, we can develop and design drugs that mirror physiologic conditions and identify natural biomolecules that can curb antimicrobial growth, one such technology being the I-chip technology (isolation chip) which employs techniques to culture the bacteria in their natural habitats meant for those bacteria that could not have been successfully cultured inside the lab.43
Inhibiting or destroying the compounds that empower bacteria to become drug resistant
Coating the drugs with biofilms and certain inhibitors that degrade or chelate with proteins or the enzymes that give bacteria its source of being converted to a superbug. Proper use of antiseptics, disinfectants, and hygiene practices such as regular hand washing and use of alcohol-based sanitizers have the potential to significantly reduce the spread of bacterial infections, thereby reducing reliance on antibiotics and ultimately decreasing levels of antimicrobial resistance.
Anti-virulence strategies include the use of secretion system inhibitor molecules that chelate or antagonize the virulence factors of the bacteria thereby preventing bacterial virulence. Sometimes devices such as biofilms or certain drugs can be used to modify and change the structure of antimicrobials so that compounds that degrade antimicrobials are rendered useless.
Rapid and accurate diagnosis of bacterial diseases and continuous surveillance over the population for incidences of antimicrobial resistance
Improved infrastructure and modern diagnostics are key to fighting AMR. Fund research and innovations to develop cost-effective diagnostic tools. Surveillance monitors local interventions, while developed nations can aid poorer countries in expanding laboratory facilities and vaccination programs. A global AMR surveillance database with standardized laboratory testing is crucial.
Reducing the use of antimicrobials in agriculture and animal husbandry
Regulating antimicrobial use in agriculture and allied sectors is crucial through policies tailored to local and national contexts. Minimizing unnecessary use and restricting access to highly antimicrobial-resistant drugs are necessary steps. Successful implementation requires interventions that include policy regulations, animal nutrition, and biosecurity measures. Changing the perception of livestock farmers regarding the excessive use of antibiotics is also crucial, given their long-term risks outweighing immediate benefits. Tighter control over antibiotic sales for food animals and incentivizing organic animal product production can promote positive change.44
Effective use of antiseptics and disinfectants
We can reduce the dependence upon antimicrobials by the use of antiseptics at local level to treat some minor infections and the surrounding ecosystem of bacteria can be changed by culturing other bacteria that have an antagonistic effect to the growth of pathogen in a phenomenon called as Amensalism. One such example is the use of probiotics in the diet which hinders the growth of harmful bacteria in gut.
Antimicrobial cycling
In this technique a class of antibiotics or a drug is removed from use for a limited time bound period and is later reintroduced in future so as an effort to resist the bacterial resistance towards that particular antibiotic molecule. Along with that prescribing a shorter course of antibiotics therapy can be used to thwart resistance.45
Overcoming antimicrobial resistance using electromagnetic force
Several studies have shown that use of targeted electromagnetic force to certain resistant bacteria have shown positive intended effect. Example: Nisin (an antimicrobial peptide) is commonly ineffective against gram negative bacteria. But the use of PEF (Pulsed Electromagnetic Field) with the dominant idea being increasing the permeability of the bacterial cell membrane to bactericidal molecules like nisin, has shown positive intended results by sensitizing the given gram-negative bacterial target cell towards nisin treatment. Therefore, the use of physical forces for changing the morphology of the targeted molecules towards the cell and electroporating the bacterial cell to increase the permeability and sensitivity towards antimicrobial molecules have now become the sought-after areas of focus and research to tackle the problem of AMR.46
A dedicated one health approach
The One Health Approach integrates human health, animal health, and environmental science by interlinking research and education. Its overarching goal is to promote public health and coordinated action to curb the menace of antimicrobial resistance and avert zoonotic threats. Use of dedicated tools of the WHO (World Health Organization) like the WHONET, a freely available software that evaluates the performance of veterinary services and tracks the scale and type of antimicrobial uses in animals. One health surveillance system is the need of hour and the benefits of it47 may take years to manifest in form of risk reduction due to antimicrobial resistance. The One Health approach and its global assessment along with the quadripartite organizations FAO-WHO-WOAH-UNEP (FAO – Food and Agriculture Organization of the United Nations; WOAH – World Organization of Animal Health; UNEP – United Nations Environment Programme) collective commitment would help achieve antimicrobial resistance prevention goals through awareness programs.48,49
Antibiotic stewardship
Antibiotic stewardship is a set of judicious interventions that aims to optimize clinical outcomes alongside bringing down the unintended and adverse consequences through selection of appropriate agent optimal dose and duration.50 It is also implementation of the systematic efforts to educate and persuade prescribers of antimicrobials to follow evidence-based prescribing, in order to stem antimicrobial overuse and combat antimicrobial resistance. This includes variety of methods like effective use of telemedicine as a tool to promote judicious evidence-based decision on antibiotic dispensing and help with treatment particularly in those setups where patients cannot physically visit hospitals and consult doctors. Putting newer antibiotics in shelf till need arises for their use, striking a balance between sustainable future free of superbugs and the pharma companies’ intent to maximize profits via extensive antibiotics sale. Incentives must be rolled out for pharma companies from setting up of tax deduction schemes, reducing the cost of clinical trials to eliminating all the bottlenecks in funding and acquiring infrastructure for research on the antimicrobial solutions.51 Good prescription practice amongst the physicians and various stakeholders to reap maximum benefits out of antibiotic stewardship by focusing both on effective treatment of patients as well as reducing collateral damage caused due to the use of these drugs as well as scaling down the prescription of broad-spectrum antibiotics where the pathogen is identified. There is an immediate need to root out the cognitive biases and develop a choice architecture that steers the prescribers towards rational prescribing of antibiotics.52 Fruitful implementation of antimicrobial stewardship involves the larger control over product information and financing of antimicrobials along with successful feedback loops, funding surveillance networks nationwide on cyclical collection, analysis of data for measuring their effectiveness and setting up quality indicator parameters for rational and optimal prescribing of antibiotics.53
Nano antibiotics
Use of nanoparticle coated antibiotics like metal ions of silver, gold copper over the antibiotic compounds have shown promising results in being proven as improving the efficacy of conventional antibiotics in tackling infections. One such study carried out promoted wound healing of infected wounds that were previously non responsive due to bacterial biofilm growth.54
Antibiotic adjuvants
Antibiotic adjuvants are the compounds that have the capacity to enhance the potency of antibiotics either by increasing its ability to target pathogen or by protecting itself from the deleterious effect of antimicrobial peptides that cleave antibiotics (reversing the inherent bacterial mechanism to be resistant). They work and increase their efficacy by disrupting the biofilm, enhancing oxidative stress of bacteria and inhibiting the working of bacterial efflux pumps.52 The use of permeabilizers as adjuvant has been proved to be efficient for treating resistant infections due to gram negative bacterial cells, they chelate and destabilize the outer lipopolysaccharide layer that previously render the bacteria resistant thereby increasing the penetrance of antibiotic inside the bacteria.55,56 This approach of using antibiotic adjuvants has mainly two benefits: (a) prolonging the lifespan of limited arsenal of antimicrobials we have and (b) easing the pressure for development of newer novel drugs targeting resistant bacteria. Some examples of adjuvants include polymyxins, aminoglycosides and polyamines.57
Host body immunomodulation
Use of probiotics to alter the microbial flora of the gut and to reduce the load of pathogenic bacteria there and use of microbiome transplantation are newer and unconventional treatments that are growing to be adopted vastly by the population. Along with that the use of monoclonal antibodies in successfully curing certain infections have been the highlights of this method. The monoclonal antibodies class of compounds have been touted to be future alternatives of antibiotics (to address the shortage of newer antibiotics in future).
Traditional medicines and ethnic practices lead the way in fighting AMR
Historically ever since the beginning of human civilization man has been traditionally treating diseases with the generational knowledge about plant extracts and their therapeutic potency with significant success. One such example is the using the existing knowledge from Indian traditional medicine system of “Ayurveda” which is predominantly a form of naturopathy to treat bacterial and other infectious diseases. This approach promotes cost effective sustainable farming by farmers alongside reducing dependence on antibiotics.58 This approach promotes the idea of One Health and produces antibiotic residue free milk and meat which have higher market value making it a profitable venture for farmers.59 One such example is the successful use of ethno-veterinary medicines to treat the lumpy skin disease in cattle as no specific antibiotic chemotherapy for lumpy skin disease have been specifically assigned as first line of medication for it; even indiscriminate use of higher version of antibiotics have been discouraged as they may jeopardize the innate immune system of animal body which fights the virus. Preparations of paste from the leaves of neem (Azadirachta indica) mixed with turmeric (Curcuma longa) have been successfully used to treat boils and wounds and ulcers over skin caused due to lumpy skin disease. The use of ethno-veterinary practice to cure bovine mastitis has been successfully documented wherein the clinical parameters became normal within 6 months,60 and prevention of mastitis in cattle using herbal formulation during dry period are successful examples in use of traditional knowledge to tackle this menace.61
Vaccines as an effective weapon to fight AMR
Effective vaccination policies may help in reducing the need to use antibiotics as they prevent large scale transmission of infectious disease and authorities should relentlessly work towards covering a significant human and livestock population under the protection of vaccines and work towards elimination of vaccine hesitancy amongst the masses.62 The advancement in newer vaccine development technologies gives a ray of hope in the era of rising antimicrobial resistance, newer vaccines with increased serotype coverage can reduce the induction of resistance where vaccination is limited to a fraction of pathogenic serotypes.63 Use of vaccine antibiotic combined approach and accelerating vaccine licensure process by removing bottlenecks for critically needed vaccines must be the top priority of policymakers to win the war against superbugs.64 Apart from all these techniques use of immunotherapeutic, quorum sensing inhibitors, biofilm disrupting compounds, RNA therapy and phage therapy have shown promising results in fighting this menace of antimicrobial resistance. Particularly in phage therapy genetically modified bacteriophages have been exploited as an effective instrument to treat antibacterial resistant infections.65-67
The use of antimicrobial peptides (AMPs) can be a better option to solve the current antibiotic resistance crisis. However, there is possibility of development of resistance against AMPs, which can cause cross-resistance to host AMPs.68 Cross-resistance to host AMPs might further affect the innate immune system of host and compromise its function in humans and animals.
Use of Artificial Intelligence and advanced computational biology
Artificial intelligence is the tool that learns and trains the computational models on the basis of available data to become more accurate and predict outcomes with minimal errors.69 Machine learning models can be used to predict the probability of a bacteria becoming a superbug along with predicting the pathway via which the pathogen becomes resistant.70 Employment of Al based models to classify and analyze AMR surveillance data predict future AMR crisis will reap a lot of benefits.71 AI can be employed to explore drug combinations from larger number of possible spaces accurately as use of drug combinations have increased therapeutic value along with reduced side effects. With the rapid strides that technology has been making use of Whole Genome Sequencing to sort out the potential targets within the bacterial cell to predict and design new antimicrobial peptides.71
Antimicrobial use in animals is a well-established cause of antibiotic resistance in humans. To prevent the emergence and spread of zoonotic epidemics, it is crucial to use antibiotics judiciously and establish legal frameworks for their rational use. Failure to do so could lead to mutant superbugs, posing a significant threat to global food security and healthcare infrastructure. Awareness campaigns involving school children, frontline health workers, and stakeholders are necessary to educate and disseminate information about the hazards of antimicrobial resistance. Collaborative efforts between veterinarians, livestock producers, human doctors, health authorities, and the public are crucial to counter this menace. Dwindling research and development pipeline on antibiotics and diminishing efficacy due to rising antimicrobial resistance should propel us to rethink our perception on use of antibiotics and take a collective immediate action. There needs to be greater investments on research and development by the governments along with more collaborations on sharing of surveillance data to monitor this menace of AMR. The urgent and worldwide threat of AMR demands an immediate and proactive global response. To achieve this, it is essential to provide comprehensive and compelling data to persuade decision makers and stakeholders at the national and international levels to treat AMR with the same level of importance as other public health issues.
ACKNOWLEDGMENTS
The authors would like to acknowledge the support and encouragement given by the Dean, College of Veterinary Science and Animal Husbandry, Odisha University of Agriculture and Technology (OUAT), Bhubaneswar, for writing this review article.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- Bernier SP, Surette MG. Concentration-dependent activity of antibiotics in natural environments. Front Microbiol. 2013;4:20.
Crossref - Chandy SJ, Naik GS, Balaji V, Jeyaseelan V, Thomas K, Lundborg CS. High-cost burden and health consequences of antibiotic resistance: the price to pay. J Infect Dev Ctries. 2014;8(9):1096-1102.
Crossref - World Bank. Drug-Resistant Infections: A Threat to Our Economic Future. Washington, DC: World Bank. License: Creative Commons Attribution CC BY 3.0 IGO. 2017.
Crossref - Dixit A, Kumar N, Kumar S, Trigun V. Antimicrobial Resistance: Progress in the Decade since Emergence of New Delhi Metallo-β-Lactamase in India. Indian J Community Med. 2019;44(1):4-8.
- Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022;399(10325):629-655.
Crossref - O’Neill J. Review on antimicrobial resistance. Antimicrobial resistance: tackling a crisis for the health and wealth of nations, 2014(4).
- Palma E, Tilocca B, Roncada P. Antimicrobial Resistance in Veterinary Medicine: An Overview. Int J Mol Sci. 2020;21(6):1914.
Crossref - Nadeem SF, Gohar UF, Tahir SF, et al. Antimicrobial resistance: more than 70 years of war between humans and bacteria. Crit Rev Microbiol. 2020;46(5):578-599.
Crossref - Alaali Z, Thani BAS. Patterns of antimicrobial resistance observed in the Middle East: Environmental and health care retrospectives. Sci Total Environ. 2020;740:140089.
Crossref - Rhouma M, Madec JY, Laxminarayan R. Colistin: from the shadows to a One Health approach for addressing antimicrobial resistance. Int J Antimicrob Agents. 2023;61(2):106713.
Crossref - Li LG, Zhang T. Plasmid-mediated antibiotic resistance gene transfer under environmental stresses: Insights from laboratory-based studies. Sci Total Environ. 2023;887:163870.
Crossref - Yassine I, Rafei R, Osman M, Mallat H, Dabboussi F, Hamze M. Plasmid-mediated quinolone resistance: Mechanisms, detection, and epidemiology in the Arab countries. Infect Genet Evol. 2019;76:104020.
Crossref - Schultsz C, Geerlings S. Plasmid-mediated resistance in Enterobacteriaceae: changing landscape and implications for therapy. Drugs. 2012;72(1):1-16.
Crossref - Carattoli A. Plasmid-mediated antimicrobial resistance in Salmonella enterica. Curr Issues Mol Biol. 2003;5(4):113-122.
- DelaFuente J, Toribio-Celestino L, Santos-Lopez A, et al. Within-patient evolution of plasmid-mediated antimicrobial resistance. Nat Ecol Evol. 2022;6(12):1980-1991.
Crossref - Guan X, Xue X, Liu Y, et al. Plasmid-mediated quinolone resistance–current knowledge and future perspectives. J Int Med Res. 2013;41(1):20-30.
Crossref - Kammili N, Rani M, Styczynski A, et al. Plasmid-mediated antibiotic resistance among uropathogens in primigravid women-Hyderabad, India. PLoS One. 2020;15(5):e0232710.
Crossref - Kivata MW, Mbuchi M, Eyase F, et al. Plasmid mediated penicillin and tetracycline resistance among Neisseria gonorrhoeae isolates from Kenya. BMC Infect Dis. 2020;20(1):703.
Crossref - Lv L, Wan M, Wang C, et al. Emergence of a Plasmid-Encoded Resistance-Nodulation-Division Efflux Pump Conferring Resistance to Multiple Drugs, Including Tigecycline, in Klebsiella pneumoniae. mBio. 2020;11(2):e02930-19.
Crossref - Seethalakshmi PS, Prabhakaran A, Kiran GS, Selvin J. Genomic insights into plasmid-mediated antimicrobial resistance in the bacterium Bhargavaea beijingensis strain PS04. Arch Microbiol. 2023;206(1):33.
Crossref - Schwarz S, Shen J, Wendlandt S, et al. Plasmid-Mediated Antimicrobial Resistance in Staphylococci and Other Firmicutes. Microbiol Spectr. 2014;2(6).
Crossref - Lahiri D, Nag M, Ghosh A, et al. Biofilm and Antimicrobial Resistance. Biofilm-Mediated Diseases: Causes and Controls. 2021:183-208.
Crossref - Datta S, Nag S, Roy DN. Biofilm-producing antibiotic-resistant bacteria in Indian patients: a comprehensive review. Curr Med Res Opin. 2024:1-20.
Crossref - Upadhyay A, Pal D, Kumar A. Interrogating Salmonella Typhi biofilm formation and dynamics to understand antimicrobial resistance. Life Sci. 2024;339:122418.
Crossref - Oschmann-Kadenbach AM, Schaudinn C, Borst L, et al. Impact of Mycobacteroides abscessus colony morphology on biofilm formation and antimicrobial resistance. Int J Med Microbiol. 2024;314:151603.
Crossref - Gattu R, Ramesh SS, Ramesh S. Role of small molecules and nanoparticles in effective inhibition of microbial biofilms: A ray of hope in combating microbial resistance. Microb Pathog. 2024;188:106543.
Crossref - Pauze-Foixet J, Mathieu-Denoncourt A, Duperthuy M. Elevated concentrations of polymyxin B elicit a biofilm-specific resistance mechanism in Vibrio cholerae. Res Microbiol. 2024:104179.
Crossref - Boroujeni MB, Mohebi S, Malekian A, et al. The therapeutic effect of engineered phage, derived protein and enzymes against superbug bacteria. Biotechnol Bioeng. 2024;121(1):82-99.
Crossref - Abebe GM. The Role of Bacterial Biofilm in Antibiotic Resistance and Food Contamination. Int J Microbiol. 2020;1705814.
Crossref - Zhao X, Yu Z, Ding T. Quorum-Sensing Regulation of Antimicrobial Resistance in Bacteria. Microorganisms. 2020;8(3):425.
Crossref - Erasmus V, Daha TJ, Brug H, et al. Systematic review of studies on compliance with hand hygiene guidelines in hospital care. Infect Control Hosp Epidemiol 2010;31(3):283-294.
Crossref - Tripathi V, Cytryn E. Impact of anthropogenic activities on the dissemination of antibiotic resistance across ecological boundaries. Essays Biochem. 2017;61(1):11-21.
Crossref - Foyle L, Burnett M, Creaser A, et al. Prevalence and distribution of antimicrobial resistance in effluent wastewater from animal slaughter facilities: A systematic review. Environ Pollut. 2023;318:120848.
Crossref - Balbin MM, Hull D, Guest C, et al. Antimicrobial resistance and virulence factors profile of Salmonella spp. and Escherichia coli isolated from different environments exposed to anthropogenic activity. J Glob Antimicrob Resist. 2020;22:578-583.
Crossref - Navarro A, Sanseverino I, Cappelli F, et al. Study of antibiotic resistance in freshwater ecosystems with low anthropogenic impact. Sci Total Environ. 2023;857(Pt 3):159378.
Crossref - Pereira SG, Alarico S, Tiago I, et al. Studies of antimicrobial resistance in rare mycobacteria from a nosocomial environment. BMC Microbiol. 2019;19(1):62.
Crossref - Swift BMC, Bennett M, Waller K, et al. Anthropogenic environmental drivers of antimicrobial resistance in wildlife. Sci Total Environ. 2019;649:12-20.
Crossref - Marston HD, Dixon DM, Knisely JM, Palmore TN, Fauci AS. Antimicrobial Resistance. JAMA. 2016;316(11):1193-1204.
Crossref - Ryan M. Assessing national action plans on antimicrobial resistance in animal production: What lessons can be drawn? OECD Food, Agriculture and Fisheries Papers. 2021;134:1-28.
Crossref - Anthony WE, Burnham CD, Dantas G, Kwon JH. The Gut Microbiome as a Reservoir for Antimicrobial Resistance. J Infect Dis. 2021;223(12 Suppl 2):S209-S213.
Crossref - Sorbara MT, Pamer EG. Interbacterial mechanisms of colonization resistance and the strategies pathogens use to overcome them. Mucosal Immunol. 2019; 12(1):1-9.
Crossref - Iacob S, Iacob DG. Infectious threats, the intestinal barrier, and its trojan horse: dysbiosis. Front Microbiol. 2019;10:1676.
Crossref - Kollef MH. Is antibiotic cycling the answer to preventing the emergence of bacterial resistance in the intensive care unit? Clin Infect Dis. 2006;43(Suppl 2):S82-S88.
Crossref - Novickij V, Staneviciene R, Vepstaite-Monstavice I, et al. Overcoming Antimicrobial Resistance in Bacteria Using Bioactive Magnetic Nanoparticles and Pulsed Electromagnetic Fields. Front Microbiol. 2018;8.
Crossref - Queenan K, Hasler B, Rushton J. A One Health approach to antimicrobial resistance surveillance: is there a business case for it? Int J Antimicrob Agents. 2016;48(4):422-427.
Crossref - Collignon PJ, McEwen SA. One health-its importance in helping to better control antimicrobial resistance. Trop Med Infect Dis. 2019;4(1):22.
Crossref - Schooneveld VT. Antimicrobial stewardship: attempting to preserve a strategic resource. J Community Hosp Intern Med Perspect. 2011;1(2):7209.
Crossref - Laxminarayan R, Duse A, Wattal C, et al. Antibiotic resistance-the need for global solutions. Lancet Infect Dis. 2013;13(12):1057-98.
Crossref - World Health Organization. One health joint plan of action (2022-2026): working together for the health of humans, animals, plants and the environment. 2022. https://www.who.int/publications/i/item/9789240059139.
- Charani E, Cooke J, Holmes A. Antibiotic stewardship programmes–what’s missing. J Antimicrob Chemother. 2010;65(11):2275-2277.
Crossref - Dyar OJ, Huttner B, Schouten J, Pulcini C. What is antimicrobial stewardship. Clin Microbiol Infect. 2017;23(11):793-798.
Crossref - Kumar SB, Arnipalli SR, Ziouzenkova O. Antibiotics in Food Chain: The Consequences for Antibiotic Resistance. Antibiotics. 2020;9(10):688.
Crossref - Li C, Budge LP, Driscoll CD, Willardson BM, Allman GW, Savage PB. Incremental Conversion of Outer-Membrane Permeabilizers into Potent Antibiotics for Gram-Negative Bacteria. J Am Chem Soc. 1999;121:931-40.
Crossref - Kwon DH, Lu CD. Polyamines increase antibiotic susceptibility in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2006;50(5):1623-1627.
Crossref - Gonzalez-Bello C. Antibiotic adjuvants – A strategy to unlock bacterial resistance to antibiotics. Bioorg Med Chem Lett. 2017;27(18):4221-4228.
Crossref - Nair MNB, Punniamurthy N. Ethno-Veterinary Practices (EVP) as a New Approach for Management of Cattle Health without Antimicrobial and Other Chemical Veterinary Drugs. EC Vet Sci. 2021;6(5):22-25
- Pradhan S and Mishra S, Ethnoveterinary practice: An alternative treatment approach in contemporary India. Pharma Innovation. 2018;7(9):362-365.
- Nair BMN, Punniamurthy N, Mekala P, Ramakrishnan N, Kumar SK. Ethnoveterinary formulation for treatment of bovine mastitis. Res Rev: J Vet Sci. 2017;18(S1):377-382.
- Kumar SK, Deepa PM, Punnimurthy N. Study on the Prevention of Mastitis in Cattle during Dry Period Using Herbal Formulation. Research Aspects in Agriculture and Veterinary Science. 2021;4:1-5.
Crossref - Michael CA, Dominey-Howes D, Labbate M. The antimicrobial resistance crisis: causes, consequences, and management. Front Public Health. 2014;2:145.
Crossref - Mitchell PK, Lipsitch M, Hanage WP. Carriage burden, multiple colonization and antibiotic pressure promote emergence of resistant vaccine escape pneumococci. Philos Trans R Soc Lond B Biol Sci. 2015;370(1670):20140342.
Crossref - Buchy P, Ascioglu S, Buisson Y, et al. Impact of vaccines on antimicrobial resistance. Int J Infect Dis. 2020;90:188-196.
Crossref - Westwater C, Kasman LM, Schofield DA, et al. Use of genetically engineered phage to deliver antimicrobial agents to bacteria: an alternative therapy for treatment of bacterial infections. Antimicrob Agents Chemother. 2003;47(4):1301-1307.
Crossref - Thiel K. Old dogma, new tricks-21st century phage therapy. Nat Biotechnol. 2004;22(1):31-36.
Crossref - Lu TK, Collins JJ. Engineered bacteriophage targeting gene networks as adjuvants for antibiotic therapy. Proc Natl Acad Sci. 2009;106(12):4629-4634.
Crossref - Roscher R, Bohn B, Duarte MF, Garcke J. Explainable machine learning for scientific insights and discoveries. IEEE Access. 2020;8:42200-42216.
Crossref - Rabaan AA, Alhumaid S, Mutair AA, et al. Application of Artificial Intelligence in Combating High Antimicrobial Resistance Rates. Antibiotics.2022;11(6):784.
Crossref - Jangir PK, Ogunlana L, Szili P, et al. The evolution of colistin resistance increases bacterial resistance to host antimicrobial peptides and virulence. Elife. 2023;12:e84395.
Crossref - Lv J, Deng S, Zhang L. A review of artificial intelligence applications for antimicrobial resistance. Biosafety and Health. 2021;3(1):22-31.
Crossref - Das P, Delost MD, Qureshi MH, Smith DT, Njardarson JT. A survey of the structures of US FDA approved combination drugs. J Med Chem. 2018;62(9):4265-4311.
Crossref - Sharma P, Dahiya S, Kaur P, Kapil A. Computational biology: Role and scope in taming antimicrobial resistance. Indian J Med Microbiol. 2023;41:33-38.
Crossref
© The Author(s) 2024. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.