ISSN: 0973-7510
E-ISSN: 2581-690X
Staphylococcus aureus causes one of the most common types of chronic mastitis. Mastitis is an inflammation of the mammary gland with local and symptoms that occasionally result in a systemic infection. This disease has profound impact on animal welfare and milk quality. This study allowed us to determine the MLST and spa typing diversity of Staphylococcus aureus isolated from cases of bovine mastitis in the west Algeria. The first purpose of this work was to investigate the phylogenetic profile of Staphylococcus aureus by spa typing and Multilocus sequence typing (MLST) isolated from Mastitis subcliniques and clinical cases which were previously investigated by Mass Spectrometry Maldi-Tof and spa typing. The second purpose was to determinate the complex clonal of strains. In total, 213 quarters milk samples were collected from 56 dairy cows at 6 farms in the region of Oran, from 2011 to 2012. The quarter milk samples were bacteriological analyzed and scored using California Mastitis Test. The prevalence-isolated bacteria were Staphylococci (38.9%)
Mastitis, Staphylococcus aureus, identification, MLST, spa, phylogeny.
Although several bacterial pathogens can cause mastitis, Staphylococcus aureus is one of the most prevalent etiologic agents of this disease in dairy cattle17, 4, Staphylococcus aureus is involved in a wide variety of diseases in humans and animals and its pathogenicity is mainly related to a combination of toxin-mediated virulence, invasive capacity, and antibiotic resistance4, 3.
As an agent of intra-mammary infections, this pathogen can contaminate the bulk milk tank and thus may constitute a bacteriological hazard for raw milk dairy products consumed. In this context, molecular subtyping tools are of great interest for the comparison of genotypes in order to identify sources and transmission routes for control improvement [5, 6], S. aureus encodes many virulence factors including the surface Ig-binding protein A (spa) whose function is to capture IgG molecules in the inverted orientation and therefore prevent phagocytosis of the bacterial cells by the host immune system11, 12, 16.
During the past decade, the epidemiology of S. aureus mastitis in dairy cattle has been studied using various molecular typing methods. MLST is a relatively new approach to identifying4, 9, MLST is an excellent technique for investigating MRSA clonality. MLST is based on the analysis of seven 500-bp sequences derived from housekeeping genes. The sequences represent distinct alleles and contain sufficient information to define the isolate using an online data search tool4, 9.
The main mechanism of methicillin resistance in S. aureus is through the expression of a foreign Penicillin binding protein (PBP), PBP2a.MRSA differ genetically from Methicillin-sensitive S. aureus isolates by the presence, in the chromosome, of a large stretch of foreign DNA (40-60 Kb), referred to as the mec element and the presence of the mecA gene that encodes the 76 KDa penicillin-binding protein PBP2a (Also referred as PBP2).
The most intensively studied species of strains of Staphylococcus, Staphylococcus aureus is a common pyogenic agent in humans and several animal species. The strains were selected as pure cultures obtained after mastitic milk cultivation on agar plates. Only one strain per case was considered for this study.
The aim of this work was to study S. aureus isolated from different cow’s milk produced in Algeria. The first aim of the present study was to use spa typing and Multilocus sequence typing (MLST) typing to investigated the phylogenetic profile of Staphylococcus aureus strains from mastitis in dairy cows, the objective was also to better determinate the clonal complex of strains. Detected MRSA isolates were further phenotypically and genomically characterized.
Bacterial strains
A selection of 56 MSSA isolates strains from cases of bovine mastitis clinical or subclinical cases of dairy bovine (n=118) in Oran were performed by bacteriological methods and by different molecular techniques (Maldi Tof and MLST, spa typing). The milk samples were collected all over Oran region during a year. The strains were selected as pure cultures obtained after mastitic milk cultivation on agar plates. Screening for pathogenic S. aureus was done by performing a determination of toxic profile by PCR in real time [4]. andvarious biochemical assays, including the coagulase test, ²-hemolysis, DNase test and growth on S. aureus agar medium. Methicillin resistance was tested on columbia agar with 5.0% salt (Oxoid) with BBL Sensi-Disc 1 ¼g Oxacillin discs (BD)4.
Mixed glycerol stocks of S. aureus cultures were prepared by suspending several loopfuls of bacteria taken by sweeping across the plate in 1.5 ml of saline with 200 ¼l of 45% glycerol for storage at “80°C. Taking a sweep across the plate rather than picking a single colony for glycerol stocks allowed us to maintain the genetic diversity of S. aureus strains in the sample for later analyses8.
Identification Maldi Tof (Matrix Assisted Laser Desorption Ionisation Time Of Flight):
The mass spectrometry allowed us to determine and confirm the identification of the species of Staphylococcus aureus strains8.
DNA extraction
To extract bacterial DNA requires the preparation of a DNA suspension from a bacterial strain8. Prepare a suspension of 4 to 5 colonies in 1 ml of sterile distilled water. Centrifuge 1 minute at 10000-12000 rpm and then remove the supernatant, Add 200 .µl of InstaGene Matrix to the pellet and incubated for 30 min at 56°C. High Speed Vortex for 10 seconds then places the tube at 100°C for 8 minutes. Collect the supernatant and store at -20°C until PCR [8].
DNA isolation was done following the mini preparation method. Isolated DNA samples were checked for purity and quantified by spectrophotometric analysis. The samples were then resolved on agarose gel (0.8%) with 1 ml of template DNA mixed with 3 ml of loading dye (xylene cyanol + bromophenol blue) and electrophoresed at 100 volts for 1 h. DNA samples showing intact bands were used for PCR amplifications.
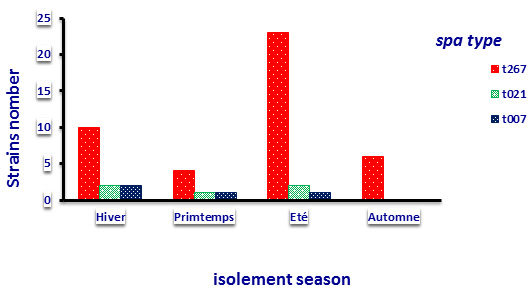 Fig. 1. Results of different types divided according to the spa isolation season during the study
Fig. 1. Results of different types divided according to the spa isolation season during the studyPCR amplifications of four virulence genes was done as described by Peacock et al14, Specific primers (Table 1) were used to amplify the four virulence genes in all S. aureus isolates. To amplify the, 25 ml of reaction mixture was made containing 20 ng of template DNA, 100 ng of primers genes, 160 mM of dNTP mix, 1.25 U Taq polymerase, 1X Taq buffer, and 0.5 mM MgCl2. All 67 isolates were amplified individually for all four genes using the specific primers, with 32 cycles of denaturation at 95°C for 1 min, annealing at 50°C for icaA, 45°C for sdrE, and 55°C for hlg and cna for 1 min, extension at 72°C for 1 min, and a final extension at 72°C for 2 min, on a DNA engine (PTC-100, MJ Research, USA). PCR products were resolved on 1.0% agarose gel at 60 volts for 2.5 h. Gels were stained with Ethidium Bromide solution (0.5 mg/ml) and documentation was done using the Gel Doc system (Bio-Rad).
Table (1):
Characteristics of 56 Staphylococcus aureus isolates recovered from bovine mastitis cases.
Strains Number |
Type mastitis case of isolation |
Spa typing |
Sequence type |
PVL positive |
MSSA |
Year of isolation |
|---|---|---|---|---|---|---|
102 |
clinical |
t 007 |
2598 |
+ |
+ |
2012 |
106 |
subclinical |
t 267 |
97 |
_ |
+ |
2012 |
111 |
subclinical |
t 021 |
39 |
_ |
+ |
2011 |
110 |
subclinical |
t 021 |
243 |
_ |
+ |
2012 |
138 |
clinical |
t 267 |
97 |
+ |
+ |
2012 |
143 |
clinical |
t 267 |
97 |
_ |
+ |
2011 |
145 |
subclinical |
t 021 |
243 |
_ |
+ |
2011 |
133 |
clinical |
t 007 |
39 |
_ |
+ |
2011 |
PVL: Planton-Valentino Leukocidin, MSSA: Methicillin sensible Staphylococcus aureus
+: Characteristics positives -: Characteristics negatives
Genotyping data production and analysis
Spa-typing
A staged spa-typing protocol was developed to enable identification of multiple-strain colonization on a large-scale. The polymorphic X region of the protein A gene (spa) was amplified with primers 1095 F: 52 -AGACGATCCTTCGGTGAGC-32 and 1517R: 52 -GCTTTTGCAATGTCATTTACTG-32 PCR reactions consisted of 0.25 mM dNTPs (Qiagen), 0.5 U of GoTaq Flexi DNA Polymerase (Promega), Colorless GoTaq Flexi Buffer, 2.5 mM of Magnesium Chloride and 0.25 ¼M of primers in a volume of 10 ¼l. PCR conditions were 94°C for 2 min, 35 cycles each of 94°C for 30 s, 50°C for 30 s, and 72°C for 60 s, and a final extension at 72°C for 5 min. PCR products were purified using Agent court AM Pure XP beads (Beckman Coulter)[8].
 Fig. 2. Revelation of spa typing on electrophoresis gel
Fig. 2. Revelation of spa typing on electrophoresis gelSamples were sequenced with the same primers as used in PCR. Sequencing reactions used BigDye v3.1 sequencing mix (Applied Biosystems) and were cycled using 30 cycles of 96°C for 10 s, 50°C for 5 s, and 60°C for 2 min. Products were purified with Agent court Clean SEQ beads (Beckman Coulter) and separated on an ABI 3730 DNA Analyzer (Applied Biosystems)[8].
Chromatograms were analyzed using Ridom Staph Type v2.0.3 software (Ridom GmbH). The relationships between spa-types were investigated using the BURP clustering algorithm incorporated into Ridom Staph Type.
Identification of rearrangements in spa-gene
A small proportion of isolates did not yield clean sequence traces with the original primers indicating the presence of rearrangements in the spa-gene. To identify possible rearrangements, primers spa-3 F: 52 -ATAGCGTGATTTTGCGGTT-32 and spa-3R: 52 -CTAAATATAAATAATGTTGTCACTTGGA-32 were used to amplify the whole spa-gene. As some isolates failed to amplify even with this extended set of primers, an alternate forward primer, spa T3-F: 52 -CAACGCAATGGTTTCATCCA-32 binding upstream from 1095 F was used together with standard reverse primer 1517R. Primer spaT3-F binds to a part of sequence encoding an IgG-binding domain of the spa-gene that is repeated five times in the gene. Due to presence of multiple binding sites for the spaT3-F primer within spa-gene, only the reverse primer (1517R) was used for sequencing10.
MLST typing
Highlighting and sequencing of a fragment of seven genes present in Staphylococcus aureus providing classification after sending the database ‘’Mlst.net’’. The seven genes are: arc (carbamate kinase), aroE (shikimate dehydrogenase ), glpF (glycerol kinase ), gmk (guanylate kinase ), pta (phosphate acetyl transferase), tpi (triosephosphate isomerase), yqiL (acetyl coenzyme A acetyl transferase). The spa tandem repeats amplification ands and condition used for MLST were performed as previously described [10]. for a subset of strains selected as representative isolates of the different spa lineages. Sequence types (STs) were assigned through the MLST database (www.mlst.net). Isolates were classified in clones by combining ST and spa lineage.
 Fig. 3. Revelation of MLST typing on electrophoresis gel
Fig. 3. Revelation of MLST typing on electrophoresis gel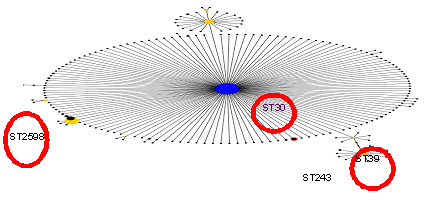 Fig. 4. Different clonal complexes of methicillin-sensitive Staphylococcus aureus those are isolated in dairy farms
Fig. 4. Different clonal complexes of methicillin-sensitive Staphylococcus aureus those are isolated in dairy farmsThe genus Staphylococcus includes several pathogenic organisms among which Staphylococcus aureus is one of the most important. The pathogenesis of staphylococcal infections is a multifactorial process that depends on expression of different virulence factors. Expression of these factors is controlled by multiple regulatory systems in conjunction with environmental signals. Most of the genetic studies in Staphylococcus aureus have been performed using different growth media, therefore, we examined the effects of different growth media on transcription of the selective target (e.g., hla, hlb, spa, sspA) and regulatory (e.g., agr, sarA family) genes.
The 56 strains were resolved into Typing MLST reveals that different spa-typing the case of isolates (S1 (t007) (st39) (TSST +) and strainS2 (T021) (pvl+) (st39)) by agains the rest of the spa-type strains (T021) have (243 st) and other strains that have proven to MLST types (st97), (st2598).
spa typing
The spa-like (RIDOM) t267 (43strains) (corresponding to the sequence of43RIDOMSRRinclassification, and the sequence [UJGFMBBBPB] in Kreis wirth classification. The spa-type (RIDOM) t021 (5strains), it corresponds to the sequence of9SRRRIDOMinclassification, and the sequence [WGKAKAOMQ] in Kreis wirth classification. The spa-type (RIDOM) t007 (4strains), it corresponds to the sequence of 10RIDOMSRR in classification, and the sequence [WGKKKKAOM] in Kreis wirth classification
The prevalence of spa-type (t267) which represents(82.69%) for the selection of S. aureus strains identified in these results are the same as obtained in America by Haran and his collaborators in 2011.
Typing protein A which showed that the strains from all dairy farms included in the study are much closed. Much of theses strains have the kind t267 (82.99%), but five t021 (9.61%) and the other four strains (7.69%) t007whichwere positive to the presence of the gene for the toxin (TSST) by detecting the toxins by real-time PCR Step One. The results obtained are comparable to those shown in the study of [14].Type distribution spa of Staphylococcus aureus strains revealed a diversity of strains of three to two types in each studied herd, as in farms F2, F3 presence of three types of strains against farms F4, F5 two types of strains of Staphylococcus aureus and one type of strain was isolated in the F6 farm.
The results of identification by MLST selected S. aureus strains shows that the strains are match able to one clonal complexand ST1: 3 with CC30 identified ST243, 39 and 2598, these strains have a derived from T021 characteristic ST30 according to the MLST database. ST97 associated with spa-type t267. The ST97 is far from being associated with theCC3O.
Some strains do not belong to ST30 frequently found in humans in Algeria, and in relation to type the ST97 strains are of bovine origin. This study is the first molecular characterization study of animal strains of S. aureus isolated in the region of Oran.
The ST-398 MRSA found in Dutch pigs has been shown not to produce the PVL toxin, but 9% of the ST-398 MRSA bacteria in humans in the Netherlands were found to have the PVL-genes. This suggests that ST-398 MRSA in pigs could still acquire this trait, or it may even suggest that some of the ST-398 MRSA bacteria in cattle or chickens are already PVL MRSA. In Korea, however, 14 MRSA samples collected from cattle over a number of years and from different regions all had the PVL gene. This particular PVL strain is believed to have developed in cattle rather than to have been acquired from humans and Ashok Srinivasan1.
Molecular typing methods are vital for the rapid identification of prevalent strains, which is important for active surveillance and to control the spread of disease. The polymorphic region of the gene encoding staphylococcal protein A (spa) has been found to be highly discriminatory and is useful for investigating both the local and global epidemiology of S. aureus.
ACKNOWLEDGMENTS
This work was financially supported by the laboratory of Unit of Microbiology, University of Rennes 1, 35043 Rennes, France and the laboratory of applied Microbiology Oran1 University Algeria. Thank you to all the teams of the microbiology unit and Renes1 University of hygiene service for their welcome and their friendship amiable. A warm thank you to Mr. Benahmed Benamar and Ms. Benahmed Rajaa
- Halpern, S.D., Ubel, P.A., Caplan, A.L. Solid-organ transplantation in HIV-infected patients. N. Engl. J. Med., 2002; 347(4): 284-7.
- Ashok, S., Matthew, J., Bankowskibc, S. E., Seifriedb, J, S., Rosalie, P., Seema, S., Yingc, C., Alan, D., Wesley, K., Randall, T. A probe-based method for confirmation of methicillin-resistant Staphylococcus aureus and detection of Panton–Valentine leukocidin and tst virulence genes. J. Diag. Infect and Dis., 2011; 70: 541-543.
- Benhamed, N., Gautier, PH., Mohamed BENKADA, M., Alioua, A., Donnio, PY., M, Kihal. Study of toxic profile of Staphylococcus aureus isolated from raw milk samples of mastitis for consumption Oran, Algeria. Inter J. Sci. Res. Sci. Technol., 2015; 6: 4-6.
- Benhamed, N ., Kihal ,M. Biodiversity of molecular profile of Staphylococcus aureus isolated from bovine mastitis cases in West Algeria. J. Bacteriol. Res., 2013; 5: 41-45.
- Benhamed, N., Moulay ,M., Aggad ,H., Henni , J. E ., ,Kihal, M. Prevalence of Mastitis Infection and Identification of Causing Bacteria in Cattle in the Oran Region West Algeria. J. Anim. Veter. Advan., 2011;10: 3002-3005.
- Bergonier, D., Sobra , A., Feßler ,T., Jacquet ,E., Gilbert, F., Schwarz ,S., Treilles, M., Bouloc, P., Pourceland, C., Vergnaud ,G. Staphylococcus aureus from 152 cases of bovine, ovine and caprine mastitis investigated by Multiple-locus variable number of tandem repeat analysis (MLVA). Vete. Res., 2014; 45:97.
- Dinesh Kumar, J, K N, Yogesh., G, Abhishek., K, Deepshikha., 2009 Detection of virulence genes in Staphylococcus aureus isolated from paper currency. Inter. J. Infect. Dis, 13. 450-455.
- Donnio, P.Y., Frederic, F., Pablo,F., Marie, B., Christele Kerve, G., Nathalie, W., Gautier, A., Lerestif, N., Lafforgue, MC. Molecular and Epidemiological Evidence for Spread of Multiresistant Methicillin-Susceptible Staphylococcus aureus Strains in Hospitals. Antimicrob. Agents and Chemoth., 2008; 51: 4342–4350.
- Enright, M. C., Spratt, B.G. Multilocus sequence typing. Trends .Microbio., 1999; l7:482-7.
- Enright, M.C., Day, N.P., Davies, C.E., Peacock, SJ., Spratt, B.G. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol., 2000; 38:1008–1015.
- Foster, T.J. Immune evasion by staphylococci. Nat. Rev. Microbiol., 2005; 3(12):948–958.
- Fournier, B., Philpott, D.J. Recognition of Staphylococcus aureus by the innate immune system. Clin. Microbiol. Rev., 2005; 18:521–540.
- Haran, K.P., Godden, S.M., Boxrud, D., Jawahir, S., Bender, J.B., Sreevatsan, S. Prevalence and characterization of Staphylococcus aureus, Including Methicillin-Resistant Staphylococcus aureus, Isolated from Bulk Tank Milk from Minnesota Dairy Farms. J. Clin. Microbiol., 2011; 50: 688-695.
- Peacock, S.J., Moore, C.E., Justice, A., Kantzanou, M., Story, l., MacKie, K., Virulent combinations of adhesion and toxin genes in natural populations of Staphylococcus aureus. Infect Immun., 2002; 70: 4987—96.
- Velusamy., S, Ashish, A., Sawant., A., Barbara, E., Gillespie, S., Headrick, J., Lorenza, C., Stephen P.Prevalence of Enterotoxin and Toxic ShocSydrome Toxin Genes in S. aureus isolated from milk of cow with Mastitis, food. Path Dis., 2006; 3:274-283.
- Votintseva, A .A., Fung, R., Ruth, R.M., Kyle ,K., Heather, G., Wyllie, D.H., Bowden, R., Derrick, W., Walker, C. Prevalence of Staphylococcus aureus protein A (spa) mutants in the community and hospitals in Oxfordshire. BMC. Microbiol., 2014; 14:63.
- Wilson, D.J, Gonzalez, R.N., Das, H.H. Bovine mastitis pathogens in New York and Pennsylvania: prevalence and effects on somatic cell count and milk production. J Dairy Sci., 1997; 80:2592–2598.
© The Author(s) 2016. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.


