ISSN: 0973-7510
E-ISSN: 2581-690X
Diarrheal diseases can lead to infections and cause morbidity and mortality in children. Diarrheagenic Escherichia coli (DEC) is an etiological agent, which is considered the major causative agent of diarrhea in children in some developing countries. The aims of this work were to estimate Escherichia coli (E. coli) causing diarrhea in children less than 5 years old, and to detect some biofilm virulence factors and the effect of some antibiotics. For the methodology, a total of 112 specimens were collected from children from two health centers, Al-Zahraa Teaching Hospital and Public Health Laboratory (located in Al-Kut city/ and the Wasit province in Iraq). All specimens were grown on simple and rich media. A total of 43 (38.4%) E. coli isolates were identified using different traditional methods, such as biochemical tests and 16S rRNA sequencing. Polymerase chain reaction (PCR) testing was used to detect some virulence factor genes that play an important role in the pathogenesis of diarrheic E. coli e.g., 16S rRNA, bfpA, and eaeA. In this study, several antibiotics were used to estimate the sensitivity and resistivity of E. coli isolates. A total of 43 isolates were fully identified as E. coli. These samples were used to detect the virulence factor genes, and 31 (72.1%) and 29 (29.4%) isolates carried bfpA and eaeA, respectively. The preponderance of E. coli isolates were completely resistant to penicillin 43 (100%). Additionally, 33 (76.7%) and 27 (62.8%) isolates were resistant to cephalothin and amoxycillin-clavulanic acid, respectively. Furthermore, the isolates of E. coli isolates showed different levels of sensitivity to antibiotics, including polymyxin B 40 (93%), norfloxacin 38 (88.4%), gentamycin 26 (60.4%), and meropenem 22 (51.2%). In conclusion, diarrheagenic E. coli isolates were the prevalent among diarrheic children. Most isolates showed varying results for the presence of virulence factors. In addition, all isolates were resistant to penicillin and sensitive to polymyxin B.
Diarrheagenic Escherichia coli (DEC), children, antibiotics
Diarrheal disease is a major global problem, causing more than 2 million deaths annually and primarily affecting children under five years of age. In addition, diarrhea is considered as one of the major disease-contributing factors for infection and death among children and the 2nd major cause of death globally among different groups of children who are under five years of age, following mortality resulting from respiratory tract infections.1,2 Hebbelstrup et al.3 mentioned that one of the major bacteria that causes diarrhea is diarrheagenic Escherichia coli (DEC), which causes gastrointestinal infections. In addition, there are six DEC pathotypes, including enteropathogenic E. coli (EPEC), enterotoxigenic E. coli (ETEC), enterohemorrhagic (Shiga toxin-producing) E. coli (EHEC STEC), enteroinvasive E. coli (EIEC), enteroaggregative E. coli (EAEC), and diffusely adhering E. coli (DAEC). ETEC, EPEC, and EAEC primarily targets the gut while DAEC, EHEC-STEC, and EIEC affect the colon. Furthermore, ETEC is a common bacterial agent that causes diarrhea and death in most developing countries. The signs of ETEC are similar to those of other bacterial infections, such as Vibrio cholera, but appear milder. The specific virulence factors resemble the enterotoxins of ETEC from other DEC.4,5 Kaper et al.5; and Alikhani et al.6 reported that EPEC is an important pathogenic group of DEC that is linked to diarrhea in children in developing countries. Strains of typical EPEC have an extra-chromosomal DNA (plasmid) named EPEC adherence factor (EAF). It encodes a type 4 pilus called the bundle-forming pilus (bfp). Several types of EPEC possess a chromosomal gene called eae gene, which encodes the ‘outer membrane protein intimin’ and affects the gastrointestinal tract mucosa. In addition, isolates of EHEC-STEC caused bloody and/or non-bloody diarrhea and hemolytic uremic syndrome. Furthermore, the virulence factor key EHEC is Shiga toxin (stx gene), which is recognized as Vero-toxin (Vtx) and consists of two subgroups: stx1 and stx2. The most important serotype among the EHEC-STEC strains were shown to be O157:H7.7 EAEC causes diarrhea in adults and travelers. This pathway is defined as a novel gut pathogen that causes various disorders worldwide. EAEC adheres to the cells of HEP2, and the mucosa of the gut by fimbria named aggregative adherence fimbria (AAFs), which is encoded by the gene (aggR), which is placed in the essential plasmids of EAEC named pAA.8,1 Jafari et al.9 clarified that EIEC strains are a patho-type that causes inflammatory invasivE. colitis, and sometimes bacterial dysentery. In most cases, EIEC causes watery diarrhea.
Bonkoungou et al.10,11 mentioned that these diseases are common in developing countries, particularly in areas with poor sanitation, hygiene, and a limited amount of safe drinking water. In addition, poor health conditions, such as malnutrition, increase the risk of infection with diarrhea. The causative agents of diarrhea, especially in acute cases, involve a wide range of pathogens, including bacteria, viruses, parasites, and fungi. In previous studies, viruses such as Rotavirus and E. coli were the two major causative agents of diarrhea, in addition to other pathogens, such as Campylobacter spp.12 Zaidi et al.13 and Vu Nguyen et al.14 demonstrated that the major causative agents of diarrhea represented by diarrheagenic pathogens include DEC, Rotavirus, bacterial dysentery (Shigella spp.), Salmonella spp., entameba dysentery (Entamoeba histolytica), Bacteroides fragilis (enterotoxigenic), Campylobacter jejuni, parasitic Cryptosporidium spp. Furthermore, DEC is a major causative agent of severe diarrhea and is a major public health.15 DEC coli is an important bacterial pathogen that causes diarrhea and death, especially during childhood age.16 In addition, DEC is a remarkable cause of childhood diarrhea and is responsible for 30–40% of acute diarrhea cases in developing countries.17 DEC is a remarkable etiological cause of both sporadic and diarrheal outbreaks worldwide. The most common DEC pathotypes cause increased morbidity and mortality globally.18
Resistance to antibiotics has recently emerged as the most common problem worldwide. This has been attributed to random sale of different antibiotics, incentives for healthcare supply to prescribe antibiotics, human expectations, and rising costs due to emergence of antibiotic resistance.19,20 Moreover, Liu et al.21 and Jones et al.22 stated that the noteworthy benefits of antibiotics in decreasing mortality and morbidity rates were challenged by the emergence of antibiotic-resistant strains in recent years. Apart from the acquisition of virulence genes by E. coli, there are a large number of cases of antibiotic resistance gene possession by the microorganism present in the clinical samples for animal and environment.23,24 Genetic changes associated with phenotypic resistance to several antibiotics, such as tetracycline, gentamycin, quinolones, sulfa-trimethoprim, and β-lactams, have been investigated in DECs.25 Jafari et al.26 demonstrated that studies in the Tehran capital (Iran) demonstrated a high frequency of resistance to STEC in populations with EAEC-, STEC-, EPEC-, and ETEC- infected children with diarrhea. In contrast, another study conducted in western Iran reported an increased phenotypic rate of resistance to EHEC in a population of STEC-, EHEC-, and EPEC- infected children.27 Bai et al.28 and Montealegre et al.29 stated that phenotype E. coli resistance is highly polymorphic, and this case is attributed to genome flexibility in E. coli, accelerating the emergence of pathogenic types showing individual resistance to the antibiotic phenotype.
The goals of this study were to estimate the incidence E. coli causing diarrhea in children less than five years old, detect some virulence factors, and determine the effect of some antibiotics.
Collect and culture for different specimens
A total of 112 clinical samples were collected from different sites, which included swabs from children’s’ stools who were hospitalized in two health centers: Al-Zahraa Teaching Hospital and the Public Health Laboratory (located in Al-Kut City, Wasit Province, Iraq). All these specimens were grown on traditional and rich media. Initially, bacteria were cultured on blood agar and nutrient agar, then on selective media, such as MacConkey, eosin methylene blue (EMB), and brain heart infusion broth (BHI B) agar; all samples were incubated 37°C for 18–20 hours. Furthermore, conventional and molecular methods have been used to extract several isolates of bacteria. Microbiological techniques, such as biochemical tests and PCR, were used to detect isolates. Moreover, Mueller-Hinton agar was used to assess antibiotic sensitivity against different E. coli isolates. Strain E-2348 was used as a control for the PCR assay (Center for Vaccine Development, USA).
Extraction protocol for E. coli DNA and PCR technique
The DNA in several isolates of E. coli were diagnosed using the Geneaid Genomic DNA Extraction Kit (U.S.A.). DNA was extracted with commensurate company guidance. Briefly, E. coli specimens were centrifuged, and the pellets were suspended in 0.2 ml buffer for ten min. A total of 0.2 ml of GD buffer was tested for ten min. Subsequently, 0.2 mL absolute of ethanol added to the lysate. A 2 ml tube was used, and then collected and centrifuged using GD columns. A buffer of W1 was added to the GD column and centrifuged. In addition, the wash buffer was tested and eluted from the column. The mixture was added and left for 3 min in order to ensure that pure DNA was obtained. Several virulence genes are required for the detection of DEC, all of which are recognized by PCR testing.
Preparation of reaction master mix for PCR
The PCR master mix was prepared using the GoTag Green Master Mix Kit (Promega, USA), and the master mix was prepared according to the manufacturer’s instructions, as summarized in Table 1.
Table (1):
Reaction of PCR mixture master mix.
Master mix of PCR |
Volume (µl) |
|---|---|
Master mix |
12.5 |
Primer forward (10p/mol) |
1 |
Primer reverse (10p/mol) |
1 |
Nuclease free water |
8.5 |
Template of DNA (20-40 ng/µl) |
2 |
Total |
25 |
Polymers chain reaction (PCR) thermo cycler program
Additionally, PCR thermocycler conditions for E. coli were achieved using a PCR thermocycler system, which is similar for each gene except for the annealing temperature, as outlined in Table 2.
Table (2):
PCR thermo cycler system of 16S rRNA, plbA, eaeA for E. coli.
| PCR Stage | Tem.(°C) | Time(m) | Repeat(cycle) |
|---|---|---|---|
| Initial Denaturation | 95 | 5 | 1 |
| Denaturation | 95 | 0.5 | 30 |
| Annealing | 531, 522, 573 | 0.5 | |
| Extension | 72 | 0.5 | |
| Final extension | 72 | 7 | 1 |
| Hold | 10 | 10 |
At for 16S rRNA1, plbA2, eaeA3
The specific primers, for example, 16S rRNA, to detect of E. coli isolates eaeA and bfpA were designed by Eurofins MWG Operon (MWG, Germany) (Table 3). The concentration and quality of the DNA specimens were estimated using a NanoDrop. The amplified DNA product was stained with ethidium bromide.
Table (3):
Primers were tested for E. coli in the present work, and selected primer to gene of 16S rRNA.
| Gene | Primer sequence | Size product (bp) | Accession No.* | |
|---|---|---|---|---|
| eaeA | F | TCCGAAAGCGAAATGATGAAG | 101 | MG458419.1 |
| R | GCCGAACCTAAGACAGGTAAG | 101 | ||
| bfpA | F | TGCTTAACACATCTGCAATTCC | 150
150 |
AF233898.1 |
| R | ATGCCGCTTTATCCAACCC | |||
| 16S rRNA | F | ACCCGCAGAAGAAAGCAAC | 230
230 |
OK177841.1 |
| R | ACGCATTTCACCGCTACAC | |||
* Designed by my self-using the national centre for biotechnology information (NCBI)
Antibiotics sensitivity test against E. coli
Sensitivity tests were conducted using the disc method, and antibiotics were selected according to the Clinical and Laboratory Standards Institute (CLSI). Mueller-Hinton agar was used for this purpose. Nutrient agar cultured with E. coli (107 CFU/ml) was incubated at 37°C for 24 hours. Discs of different antibiotics were placed on the surface of the agar. The antibiotics used in the current study were as follows: penicillin (PEN) 10U, amoxicillin-clavulanic acid (AMC) 10µg, gentamicin (GEN) 10µg, meropenem (MPM) 10µg norfloxacin (NOR) 10µg, trimethoprim-sulfamethoxazole (SXT) 250µg, cephalothin (INN) 30µg, polymyxin B (PMB) 200U. The results of this method (resistant, intermediate or susceptible) were conducted according to the CLSI system. All E. coli isolates were tested for multidrug resistant (MDR).
Statistical analysis method
All data were subjected to a one-way analysis of variance (ANOVA). We considered a P-value of 0.05 to be statistically significant.
Isolation and identification of E. coli
A total of 43 (38.4%) E. coli, was obtained from stool samples from children. These isolates were identified using conventional and molecular methods, such as culture and microscopic examination, biochemical tests, and PCR, and all results were confirmed using molecular techniques, such as 16S rRNA. All bacterial isolates demonstrated similar results across several biochemical tests. Several tests were conducted to confirm the characterization of E. coli by 16S rRNA. The DNA of all E. coli isolates was extracted, and PCR was performed. All isolates were identified as DEC.
Identification of E. coli by PCR technique
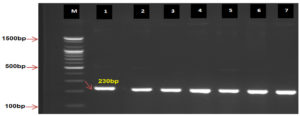
Different E. coli isolates were identified as DEC using 16S rRNA as PCR positive (Fig. 1). As for virulence factors, in DEC, bfpA was detected in 31 (72.1%) isolates from E. coli as PCR positive, whereas the eaeA gene was detected in 29(67.4%) isolates and considered a PCR positive (Fig. 2 and 3). These genes play a critical role in the pathogenesis of diarrhea caused by E. coli.
Fig. 1. Agarose gel electrophoresis and genomic DNA isolated from E. coli observed PCR product analysis for16S rRNA gene in E. coli. M: Marker (100-1500bp). All lines (1-7) were +ve PCR at 230bp PCR size of product.
Fig. 2. Agarose gel electrophoresis and genomic DNA isolated from E. coli observed PCR product analysis bfpA gene in E. coli. M: Marker (100-1500bp). All lines (1-6) were +ve PCR at 156bp PCR size of product.
Fig. 3. Agarose gel electrophoresis and genomic DNA isolated from E. coli observed PCR product analysis eaeA gene in E. coli. M: Marker (100–1500 bp). All lines (1-6) were +ve PCR at 101 bp PCR size of product.
Antibiotics sensitivity test against E. coli
In the present study, most E. coli isolates were multi drug resistant (MDR) to varying degrees (Table 3). E. coli isolates were fully resistant to penicillin (100%). A total of 33 (76.7%) and 27 (62.8%) isolates exhibited resistance to cephalothin and amoxycillin-clavulanic acid, respectively. In addition, the isolates of E. coli showed different levels of sensitivity to each antibiotic. For example, we observed sensitivity to polymyxin B 40 (93%), norfloxacin 38 (88.4%), gentamycin 26 (60.4%), and meropenem 22(51.2%), as summarized in Table 4.
Table (4):
Antibiotic resistivity and susceptibility in different isolates of DEC.
| Antibiotics dosage (µg) | Resistant | Intermediate | Sensitive | ||||
|---|---|---|---|---|---|---|---|
| No*. | (%) | No. | (%) | No. | (%) | **Total No. | |
| Penicillin G | 43 | 100 | 0 | 0 | 0 | 0 | 43 |
| Amoxicillin-Clavulanic acid | 27 | 62.8 | 7 | 16.3 | 9 | 20.9 | 43 |
| Cephalothin | 33 | 76.7 | 4 | 10.3 | 6 | 14 | 43 |
| Meropenem | 20 | 46.5 | 1 | 2.3 | 22 | 51.2 | 43 |
| Gentamicin | 14 | 32.6 | 3 | 7 | 26 | 60.4 | 43 |
| Norfloxacin | 4 | 9.3 | 1 | 2.3 | 38 | 88.4 | 43 |
| Polymyxin B | 2 | 4.7 | 1 | 2.3 | 40 | 93 | 43 |
| Trimethoprim-sulfamethoxazole | 2 | 4.7 | 39 | 90.7 | 2 | 4.7 | 43 |
(P<0.05), No*: Number, **: E. coli is MDR
Most types E. coli are harmless and cause diarrhea. Some strains E. coli (i.e. E. coli O157:H7) can cause dangerous symptoms, such as stomach cramps, vomiting, bloody, and diarrhea. Successful management of any infectious disease requires recognition of the causative agents and treatment of signs manifested by the disease. This study was conducted to detect E. coli isolation rate and virulence factors of pathogenic E. coli that isolated from DEC in the Wasit province (Iraq). Diarrhea is a multifactorial disorder related to a wide range of pathogens, including bacteria, viruses, and parasites.21,30 Most commonly isolated bacteria among DEC is E. coli when applying traditional and molecular methods. The results of the current study are in agreement with those of Begum et al.,31 conducted in Mizoram. In the current study, the prevalence of DEC was higher than that of the other microorganisms (Table 2). The current study agreed with other studies conducted in Iran/Tahran and Tanzania by Jafari et al.26, and Moyo et al.,32 respectively, who demonstrated that the most common microorganism was DEC (7.9%), which was lower than that reported in other developing countries. In addition, a study by Dias et al.33 (Brazil and Mexico) observed that EAEC was the primary pathotype DEC, with respective rates of 50%.
Regarding non-DEC and the capacity to cause diarrhea, contagious diseases, which do not first affect the GIT, can cause acute diarrhea. The pathogenesis of this type of diarrhea involves intestinal inflammation, cytokine action, red blood cell (RBC) sequestration, programmed cell death, increased endothelial cell permeability in the GIT microvasculature, and invasion of epithelial cells in the GIT by several agents. Several symptoms, such as fever and diarrhea, occur in patients with respiratory syndrome (SARS), Plasmodium parasites (malaria), and dengue fever. Diarrhea also occurs in patients with acquired lung inflammation when it is suggestive of legionellosis, and those with systemic bacterial infections. Although diarrhea is rare in patients with early borreliosis, the incidence is high in those with other tick-borne infections, such as tick-borne, ehrlichiosis, and others. Unfortunately, it is often not established whether diarrhea is an initial clinical sign and/or whether it progresses during the course of the disorder.34
Recently, molecular diagnostic techniques have become common in clinical laboratories. PCR is capable of detecting several pathogens via the amplification of specific genes encoding important virulence factors. If it is difficult to diagnose DEC using traditional laboratory techniques, PCR becomes beneficial in clinical laboratories because of its specificity and sensitivity. In the current study, all isolates produced 43 (100%) as E. coli by PCR method. In contrast, there were 31 (72.1%) and 29 (67.4%) isolates from E. coli contained the bfpA, and eaeA genes, respectively. Furthermore, typical EPEC is the most common cause of watery diarrhea in children, especially in developing countries. The current study is compatible with and nearest to a study carried out on children in Peru (South America) by Contreras et al.35, who observed that the most common pathotype in diarrhea was the predominant genes of bfpA (74%) and eaeA (54%). In addition, a study conducted in Yogyakarta/Indonesia by Harti et al.,36 who observed that the percentage of predominant genes in DEC was espA (85%), bfpA (80%), and eaeA (51%). Another study conducted among young children in South Africa with diarrheal disease, by Galane and Roux,37 clarified that the PCR results observed 59 (32.6%) isolates of E. coli carried genes (eaeA), and 6(3.3%) possessed bfpA genes, 4 (2.2%) CNF1, and 2 (1.1%) carried Stx2 genes. These results were different from those of the current study, and this difference between the two studies may be ascribed to the conditions and various geographical areas and/or other related genes or carried on plasmids, among other external causes. Moreover, Kaper et al.5 stated that the eaeA e gene is a known virulence factor not only for EPEC and EHEC strains, but is also atypical for EPEC, in which eaeA takes place alone without the presence of a gene, such as the adherence factor plasmid (pEAF).
Resistivity and antibiotic sensitivity have become important health problems, particularly in pharmacies. Numerous types of bacteria have been found to be capable of resisting antibiotics. Among these bacteria is E. coli. It is one of the microbes that cause diarrhea. Several antibiotic therapies are recommended when diarrheal diseases are present to reduce the duration of clinical signs and symptoms. E. coli in vitro isolates showed a high resistance pattern to several β-lactam antibiotics, including penicillin G 43(100%), cephalothin 33(76.7%), and amoxycillin-clavulanic acid 27(62.8%), but there was an intermediate effect on trimethoprim-sulfamethoxazole 39(90.7%). The majority of antibiotic resistance refers to the β-lactamase enzyme that destroys β-lactam antibiotics. These results agree with those of another study conducted by Wu et al.38 who observed that some antibiotics emerged as resistant to penicillin group i.e. amoxicillin (85%), followed by cephoms group, such as cefuroxime (65%), and cefatriaxone (60%), respectively. Furthermore, it was found in the current study that these results were compatible with the results of a study conducted in Katsina State, Nigeria by Adesoji and Liadi,39 who observed that isolates of E. coli demonstrate high resistance to several antibiotics, including ampicillin (100%), amoxicillin (80%), and tetracycline (73.3%). Different cases of diarrhea caused by various categories of E. coli constitute risk in several regions of the world.40 There are studies conducted in the Ondo state/ Nigeria by Onifade et al.,41 who observe that most E. coli isolates are resistant to antibiotics (i.e. ceftazidime, augmentin, gentamicin, cefuroxime, tetracycline, cefixime, trimethoprim-sulfamethoxazole, and chloramphenicol) but have observed >50% susceptibility to ofloxacin, ciprofloxacin, and nitrofurantoin. The resistance rate E. coli to drugs has been observed to be different from one location to another in Nigeria. Antibiotics such as CHL, SXT, and AMP are excessively used to treat diarrhea owing to their low cost and ready availability. Some prior studies have observed a high prevalence of antibiotic resistance in pathogens, especially DEC. Furthermore, the present study has exhibited prevalece sensitivity to some antibiotic, such as polymyxine B (93%), norfloxacin (88.4%), and gentamicin (60%). These results agreed with those of a study conducted in Iran by Broujerdi et al.,43 who reported that most E. coli isolates (> 75%) were sensitive to some antibiotic groups, such as imipenem, ciprofloxacin, and amikacin.
Infection with diarrheic E. coli is the most common pathogen among microbial communities and remains as one of the primary causes of childhood diarrhea. In the present study, the case of DEC, particularly EPEC, was considerably associated with childhood diarrhea in developing countries. Furthermore, molecular techniques have indicated that bfpA and eaeA genes are associated with the form of DEC in different isolates. The results of the antibiotic susceptibility assay indicated that the most active compound against DEC inhibited different DEC isolates. Moreover, isolates of E. coli are fully resistant to penicillin (100%). In addition, the isolates of E. coli demonstrated sensitivity to polymyxin B (93%) and norfloxacin (88.4%).
ACKNOWLEDGMENTS
The authors would like to thank the writing support staff at the centre of Al-Zahra’ Teaching Hospital and the public Health Laboratory in Al-Kut city/Wasit province/Iraq
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
- Kosek M, Bern C, Guerrant RL: The global burden of diarrhoeal disease, as estimated from studies published between 1992 and 2000. Bull World Health Organ. 2003;81:197-204.
- Bryce J, Boschi-Pinto C, Shibuya K, Back RE: WHO estimates of the causes of death in children. Lancet. 2005;365(9465):1147-1152.
Crossref - Hebbelstrup Jensen B, Olsen KEP, Struve C, Krogfelt KA, Petersen AM. Epidemiology and clinical manifestations of enteroaggregative Escherichia coli. Clin Microbiol Rev. 2014;27(3):614-630.
Crossref - Fleckenstein JM, Rasko DA. Overcoming enterotoxigenic E. coli pathogen diversity: Translational molecular approaches to inform vaccine design. Methods Mol Biol. 2016;1403:363-383.
Crossref - Kaper JB, Nataro JP, Mobley HLT. Pathogenic E. coli. Nat Rev Microbiol. 2004;2:123-140.
Crossref - Alikhani MY, Hashemi SH, Aslani MM, Farajnia S. Prevalence and antibiotic resistance patterns of diarrheagenic E. coli isolated from adolescents and adults in Hamedan, Western Iran. Iran J Microbiol 2013;5(1):42-47.
- Luzader DH, Willsey GG, Wargo MJ, Kendall MM. The type three secretion system 2-encoded regulator EtrB modulates enterohemorrhagic E. coli virulence gene expression. Infect Immun. 2016;84(9):2555-2565.
Crossref - Boisen N, Struve C, Scheutz F, Krogfelt KA, Nataro JP. New adhesin of enteroaggregative E. coli related to the Afa/Dr/AAF family. Infect Immun. 2008;76(7):3281-3292.
Crossref - Jafari A, Aslani MM, Bouzari S. E. coli: a brief review of diarrheagenic pathotypes and their role in diarrheal diseases in Iran. Iran J Microbiol. 2012;4:102-117.
- Bonkoungou IJ, Sanou I, Bon F, et al. Epidemiology of rotavirus infection among young children with acute diarrhoea in Burkina Faso. BMC Pediatr. 2010;10:94.
Crossref - Bonkoungou IJ, Lienemann T, Martikainen O, et al. Diarrhoeagenic Escherichia coli detected by 16-plex PCR in children with and without diarrhoea in Burkina Faso. Clin Microbiol Infect. 2012;18(9):901-906.
Crossref - Scallan E, Hoekstra RM, Angulo FJ, et al. Foodborne Illness acquired in United states-major pathogens. Emerg Infect Dis. 2011;17(1):7-15.
Crossref - Zaidi AKM, Awasthi S, DeSilva HJ. Burden of infectious diseases in South Asia. BMJ. 2004;328:811-815.
Crossref - Nguyen T, Le Van P, Le Huy C, Nguyen Gia K, Weintraub A. Etiology and epidemiology of diarrhoea in children in Hanoi, Vietnam. Int J Infect Dis. 2006;10(4):298-308.
Crossref - Munhoz DD, Santos FF, Mitsunari T, et al. Hybrid Atypical Enteropathogenic and Extraintestinal Escherichia coli (aEPEC/ExPEC) BA1250 Strain: A Draft Genome. Pathogens. 2021;10(4):475.
Crossref - Ugboko HU, Nwinyi OC, Oranusi SU, Oyewale JO. Childhood diarrhoeal diseases in developing countries. Heliyon. 2020;6(4):e03690.
Crossref - Saka HK, Dabo NT, Muhammad B, Garcia-Soto S, Ugarte-Ruiz M, Alvarez J. Diarrheagenic Escherichia coli Pathotypes From Children Younger Than 5 Years in Kano State, Nigeria. Front Public Health. 2019;27(7):348.
Crossref - Croxen MA, Law RJ, Scholz R, Keeney KM, Wlodarska M, Finlay BB. Recent advances in understanding enteric pathogenic Escherichia coli. Clin Microbiol Rev. 2013;26(4):822-880.
Crossref - Laxminarayan R, Chaudhury RR. Antibiotic resistance in India: drivers and opportunities for action. PLoS Med. 2016;13(3):e1001974.
Crossref - Kakkar M, Walia K, Vong S, Chatterjee P, Sharma A. Antibiotic resistance and its containment in India. BMJ. 2017;358:j2687.
Crossref - Liu L, Johnson HL, Cousens S, et al. Global, regional, and national causes of child mortality: an updated systematic analysis for 2010 with time trends since 2000. Lancet. 2012;379(9832):2151-2161.
Crossref - Jones RN, Sader HS, Fritsche TR, Pottumarthy S. Comparisons of parenteral broad-spectrum cephalosporins tested against bacterial isolates from pediatric patients: report from the SENTRY antimicrobial surveillance program (1998-2004). Diagn Microbiol Infect Dis. 2007;57(1):109-116.
Crossref - Van TTH, Chin J, Chapman T, Tran LT, Coloe PJ. Safety of raw meat and shellfish in Vietnam: an analysis of Escherichia coli isolations for antibiotic resistance and virulence genes. Int J Food Microbiol. 2008;124(3):217-223.
Crossref - Titilawo Y, Obi L, Okoh A. Antimicrobial resistance determinants of Escherichia coli isolates recovered from some rivers in Osun state, South-Western Nigeria: implications for public health. Science of the Total Environment. 2015;523:82-94.
Crossref - Kipkorir KC, Ang’ienda PO, Onyango DM, Onyango PO. Antibiotic Resistance of Escherichia coli from Humans and Black Rhinoceroses in Kenya. Ecohealth. 2020;17(1):41-51.
Crossref - Jafari F, Garcia-Gil LJ, Salmanzadeh-Ahrabi S, et al. Diagnosis and prevalence of enteropathogenic bacteria in children less than 5 years of age with acute diarrhea in Tehran children’s hospitals. J Infect. 2009;58(1):21-27.
Crossref - Bouzari S, Farhang E, Hosseini SM, Alikhani MY. Prevalence and antimicrobial resistance of shiga toxin-producing Escherichia coli and enteropathogenic Escherichia coli isolated from patients with acute diarrhea. Iran J Microbiol. 2018;10(3):151-157.
- Bai X, Zhang J, Ambikan A, et al. Molecular Characterization and Comparative Genomics of Clinical Hybrid Shiga Toxin-Producing and Enterotoxigenic Escherichia coli (STEC/ETEC) Strains in Sweden. Sci Rep. 2019;9(1):5619.
Crossref - Montealegre MC, Talavera Rodriguez A, Roy S, et al. High Genomic Diversity and Heterogenous Origins of Pathogenic and Antibiotic-Resistant Escherichia coli in Household Settings Represent a Challenge to Reducing Transmission in Low-Income Settings. mSphere. 202015;5(1):e00704-e00719.
Crossref - Saeed A, Abd H, Sandstrom G. Microbial aetiology of acute diarrhoea in children under five years of age in Khartoum, Sudan. J Med Microbiol. 2015;64:432-437.
Crossref - Begum J, Tapan B, Choudhary PR. Antimicrobial assay of shigatoxigenic E. Coli (STEC) and enteropathogenic E. coli (EPEC) isolated from diarrhoeic faecal samples of piglets and infants in Mizoram. Indian Journal of Animal Sciences. 2015;85(10):1067-1072.
- Moyo SJ, Gro N, Matee MI, et al. Age specific aetiological agents of diarrhoea in hospitalized children aged less than five years in Dar es Salaam, Tanzania. BMC Pediatr. 2011;11:19.
Crossref - Dias RC, Dos Santos BC, Dos Santos LF, et al. Diarrheagenic Escherichia Coli pathotypes investigation revealed atypical enteropathogenic E. Coli as putative emerging diarrheal agents in children living in Botucatu, Sao Paulo state, Brazil. APMIS. 2016;124(4):299-308.
Crossref - Reisinger EC, Fritzsche C, Krause R, Krejs GJ. Diarrhea caused by primarily non-gastrointestinal infections. Nature Clinical Practice Gastroenterology & Hepatology. 2005;2(5):216-222.
Crossref - Contreras CA, Ochoa TJ, Lacher DW, et al. Allelic variability of critical virulence genes (eae, bfpA and perA) in typical and atypical enteropathogenic Escherichia coli in Peruvian children. J Med Microbiol. 2010;59(1):25-31.
Crossref - Harti S, Iravati S, Asmara W. Detection of eae, bfpA, espA Genes on Diarrhoeagenic Strains of Escherichia coli Isolates. Indonesian Journal of Biotechnology. 2006;11(1):889-889
Crossref - Galane PM, Roux ML. Molecular epidemiology of Escherichia coli isolated from young South African children with diarrhoeal diseases. J Health Popul Nutr. 2001;19(1):31-38.
- Wu D, Ding Y, Yao K, Gao W, Wang Y. Antimicrobial Resistance Analysis of Clinical Escherichia coli Isolates in Neonatal Ward. Front Pediatr. 2021;25(9):670470.
Crossref - Adesoji AT, Liadi AM. Antibiogram studies of Escherichia coli and Salmonella species isolated from diarrheal patients attending Malam Mande General Hospital Dutsin-Ma, Katsina State, Nigeria. Pan Afr Med J. 2020;37:110.
Crossref - Joffre E, Rojas VI. Molecular Epidemiology of Enteroaggregative Escherichia coli (EAEC) Isolates of Hospitalized Children from Bolivia Reveal High Heterogeneity and Multidrug-Resistance. IJMS. 2020;21(24).
Crossref - Onifade AK, Oladoja MA, Fadipe DO. Antibiotics sensitivity pattern of E. coli isolated from children of school age in Ondo state, Nigeria. Researcher. 2015;7(2):73-76.
- Broujerdi SM, Ardakani MR, Rezatofighi SE. Characterization of diarrheagenic Escherichia coli strains associated with diarrhoea in children, Khouzestan, Iran. J Infect Dev Ctries. 2018;12(8):649-656.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.