ISSN: 0973-7510
E-ISSN: 2581-690X
Streptococcus pyogenes was the most common bacterial causes of tonsillitis. The detection of virulence factors of this pathogen can be used to determine pathogenic potential as a rapid screening method. A total of 109 isolates (46%) showed positive culture for S. pyogenes, these isolates recovered from 235 swabs were collected from tonsillitis patients in ENT department in Al–Habboby Teaching Hospital, Thi-Qar province, Iraq. S. pyogenes isolates exposed to detect the specific gene (spy1258)and one of the virulence factors were smeZ gene by PCR technique and DNA sequencing analysis. The PCR results recorded that 61% and 50% of isolates harbor spy1258 and smeZ genes, respectively. The sequencing of PCR products showed significant alignments identities (94-100%) to the S. pyogenes for both genes which are located in BLAST-NCBI Genbank. The four PCR products of both genes were registered in Genbank under the named as (ZKD1 Spy-like gene; ZKD2 Spy-like gene; ZKD3 SmeZ-like gene and ZKD4 SmeZ-like gene). The results of Multiple sequence alignment analysis recorded that C > T and T > C polymorphism for smeZ gene.
S. pyogenes, tonsillitis, spy1258, smeZ, gene sequences.
Group A streptococcus, S. pyogenes (GAS) was a common human bacteria responsible for a wide range of diseases, from restricted throat infections like pharyngitis, tonsillitis, to hostile infections as streptococcal toxic shock syndrome, sepsis, bacteraemia, and necrotizing fasciitis1. Even though’ the S. pyogenes was repeatedly present extra cellulary in the host cells. Hertze et al.,2 demonstrated that this bacteria might exist in intracellularly and evade the killing by in cooperation the human macrophages and /or neutrophils3 .
The austerity of GAS infections were hinged on various factors of bacteria and host. Also, the pathogenic possessions of S. pyogenes isolates were allied to creation of the multiple virulence factors like DNases, proteases, toxins, and other toxins which present in goal bacteria (Group A streptococcus) might be the forecaster of its invasiveness4 . Huge group of toxins coded by smeZ, speM, speC, speIgenes is involved in systemic toxicity5 .
The complex pathogenicity of GAS were related to its ability to production of numerous virulence factors such as Streptococcal mitogenic exotoxin Z (smeZ) was stimulated the immune system, and its linked with different disease like scarlet fever, acute rheumatic fever, and toxic shock syndrome6.
‘The polymerase chain reaction (PCR) technique directing to transcriptional regulator genes supplied the rapid and dependable manner for detection of a pathogenic microbes7. The transcriptional regulators were dedicated a DNA binding proteins that performance a critical role in guiding the gene expression inside microbes aimed at the adaptation and endurance in diverse environment. A spy1258 gene was one of reputed transcriptional regulator gene (TetR/AcrR family) that was precise for GAS and could be used as a marker for the detection of this bacteria8 .
Liu et al.,8 described spy1258 gene that is exceptionally lay out in isolates of S. pyogenes. The using of PCR technique of spy1258 gene assisted an amplification of DNA that extracted from GAS only, but never from the other Streptococcus species and public bacteria. The main goal of this study was to detect the GAS, to establish the sensitivity of spy1258 gene to identification of S. pyogenes, to determine the occurrence of smeZ gene in this bacteria. Furthermore, to comparative genomic sequencing analysis permits for an epidemiological investigation of closely associated bacterial isolates.
Ethical approval
This research was approved by the Medicine College Ethics Committee, Thi-Qar University, Thi-Qar Province, Iraq.
Bacterial isolates
Two hundred and thirty five swabs were collected from patients infected with tonsillitis whom admitted to Ear Nose Throat (ENT) unit in AL-Habbuby Teaching Hospital of Thi-Qar province, during the period from November, 2015to May, 2016 by moistened sterile swabs with normal saline, these swabs directly inoculated on bloodagar (LAB/ United Kingdom) and incubated at 37°C for 24 hour.
Identification of S. pyogenes
- pyogenes was identified depending on morphological properties on culture media, biochemical tests and Bacitracin susceptibility using 0.04 units Bacitracin discs (Bio analyse/Turkey) 9,10. S. pyogenes diagnosis was confirmed by API system (BioMerieux / France). Lastly, to serological identification of S. pyogenes by used the Mast Streptococcus kit (Mast/United Kingdom), it is a rapid latex slide agglutination for identification of Streptococci of ‘Lancefield groups A,B,C,F and G, according to the instructions of the manufactured company.
Preparation of bacterial DNA
Completely S. pyogenes isolates were inoculated on Brain Heart Infusion broth (LAB/United Kingdom) and incubated for 24h at 37°C. ‘The DNA of bacteria extracted from a fresh culture in a Brain Heart Infusion broth by using DNA Bacteria plus kit (Geneaid / Korea) according to the manufacturers instructions’.
Polymerase Chain Reaction diagnosis of spy1258 and smeZ genes
The specific primer pairs of spy1258 gene as following: forward : 5’-AAAGACCG CCTTAACCACCT-3’ and reverse: 5’-TGGCAAGGTAAA CTTCTAAAGCA-3’8, whereas for smeZ gene: forward: ‘5’-TTTCTCGTCCTGTGTTTGGA-3’ and reverse : 5’ TTCCAATCAAATGGGACGGAGAACA-3’.
The PCR cycling program of spy1258 gene: initial denaturation at 94°C for 2 minutes, followed by 30 cycles of denaturation at 94°C for 20 seconds, annealing at 55°C for 20 seconds, extension at 72°C for 45 seconds and final extension for 2 minutes after the last cycle11 , while for smeZ gene was set at 95°C for 5 minutes followed by 30cycles of 94°C for 30 second, 58°C for 30seconds, and 72°C for 1 minute, and final extension at 72°C for 5 minutes’12 .
The visualization of PCR products was showed in 2% agarose gelelectrophoresis and the attendance of a 407bp and 246bp band as a positive result for spy1258 and smeZ genes, respectively.
DNA sequencing
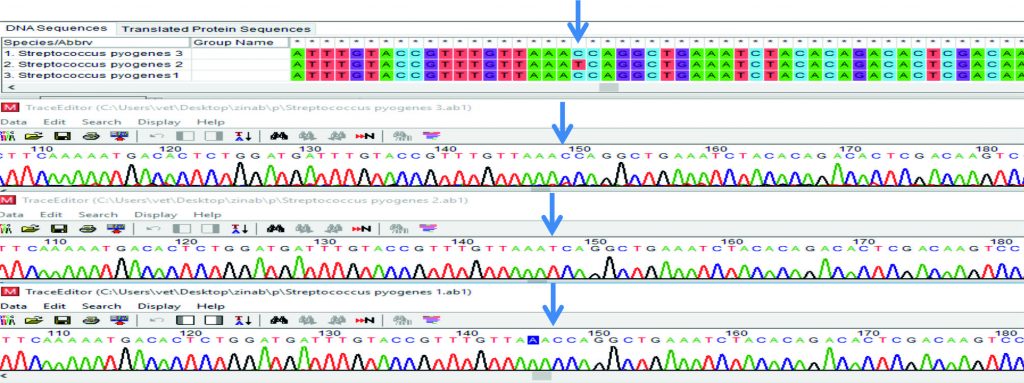
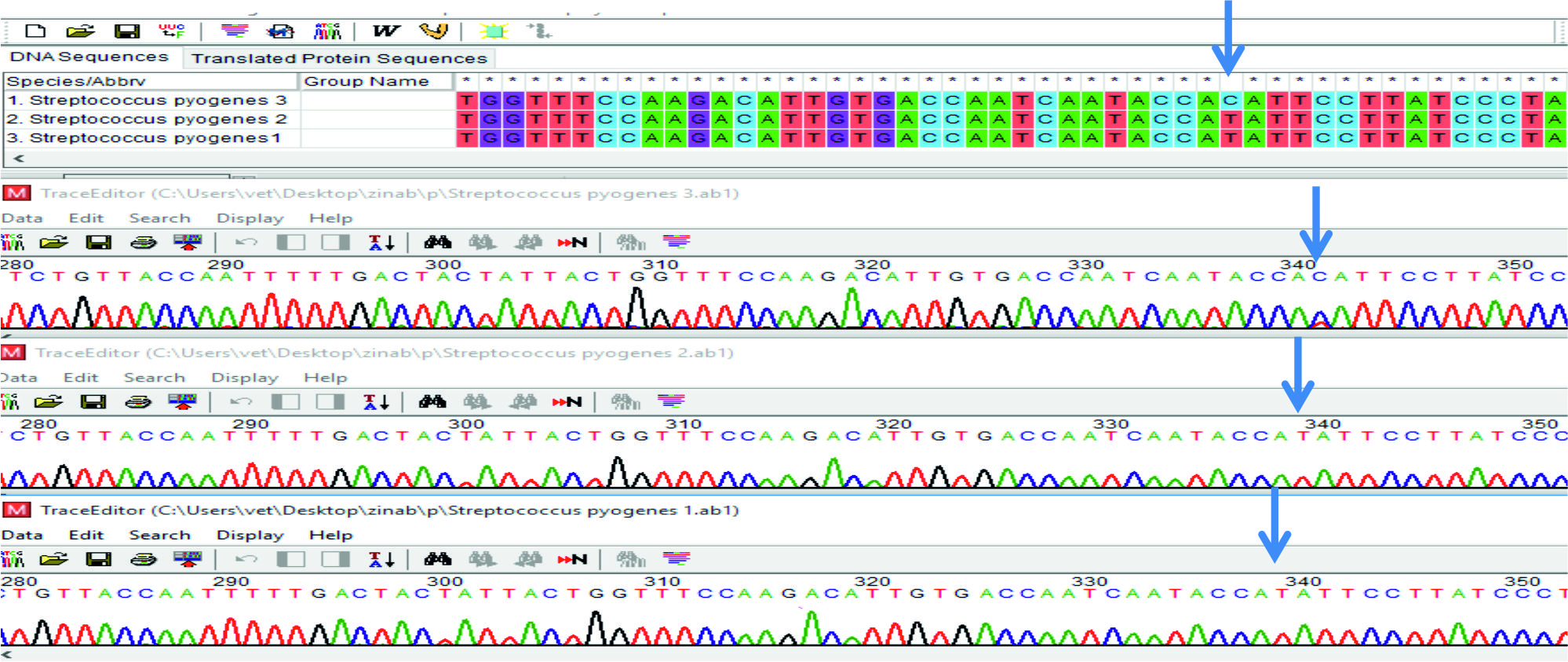
Seven PCR products of S. pyogenes distributed to three for spy1258 and four to smeZ genes, were selected for sequencing and forward and reverse primers for each gene were sent to the laboratory to be sequenced (Macrogen, Korea). Basic Local Alignment Search Tool analysis (BLAST) was lead to blast algorithm (www.ncbi.nlm.nih.gov/ BLAST). The sample sequences designated as (ZKD1, ZKD2, ZKD3, ZKD4, ZKD5, ZKD6 and ZKD7) for both genes were edited, aligned, and compared with the reference sequences using Bio Edit sequence Alignment Editor Software Version 7.1 (DNASTAR, USA)13 (Hall, 1999). A phylogenetic tree for each gene sequence was constructed by using MEGA7 software14.
The results of the present study displayed that the occurrence of S. pyogenes was 109/235 isolates (46%), S. pyogenes was one of recurrent bacterial agent of tonsillitis. Simon,15 ; and Beye et al.,16 recorded that S. pyogenes was an important pathogen causes of tonsillitis, cutaneous and systemic infections.
The current results incorporated with results of 17 showed that S. pyogenes and S. aureus were caused tonsillitis and pharyngitis and revealed 35.42% as positive culture for S. pyogenes, while the frequency of S. pyogenes was higher than the results of18 recorded that the most agents of tonsillitis were S. pyogenes with a percentages of 20.2 %.
The frequency of spy1258 gene in S. pyogenes isolates was 61%, and the size of this gene was approximately 407 bp, Fig. (1). spy1258 was definite gene only for GAS and used for identification of this species. Dunne et al.,19 revealed that the spy1258 gene was the major gene targets of S. pyogenes. Also7 recorded that this gene was specific ‘for GAS and could be involved in species to specific conservation or adaptation’. On other hand, the current results agreed with studies performed by8 and20 , those studies showed that spy1258 gene was specific for S. pyogenes only, but not from other species of the genus Streptococcus and common bacteria.
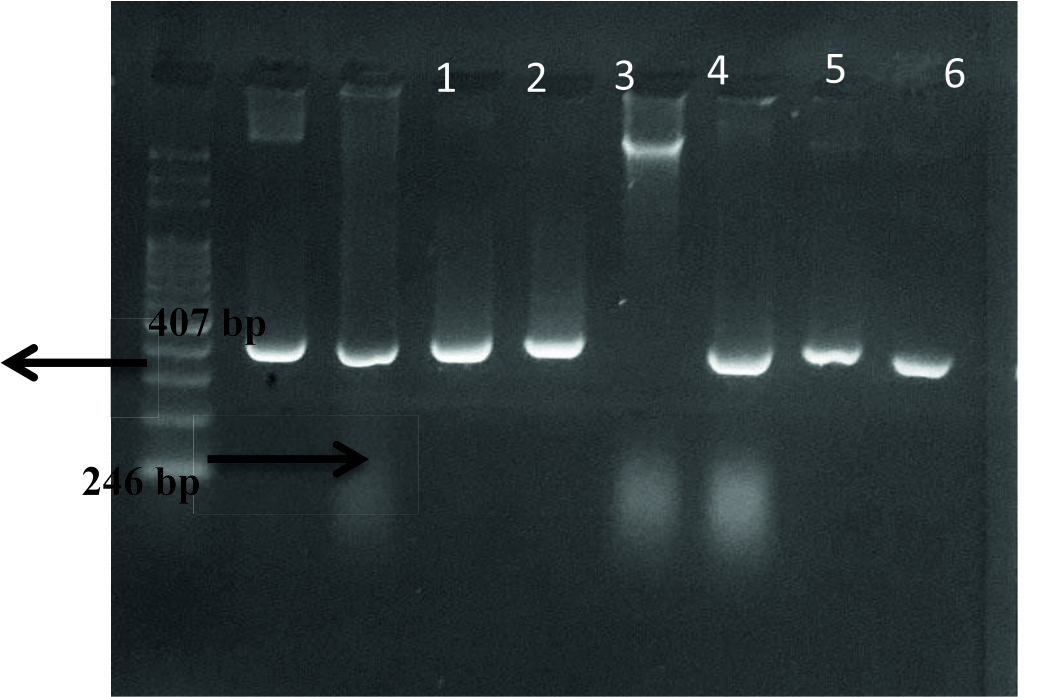
Fig. 1. Agarose gel electrophoresis of spy1258gene amplification,M: ladder, 1-4, 6-8: positive results, 5: negative results.
The present study was disagreed with results of21,22 displayed that all S.pyogenes isolates contains spy1258 gene which isolated from patients with tonsillitis, while19 recorded that 21 isolates from 24 S. pyogenes had this gene, also the spy1258 gene had lesser sensitivity to detection three isolates of S. pyogenes.
The existence of smeZ gene was 50% in S. pyogenes isolates, Fig. (2) showed the size of this gene which was approximately 246 bp. smeZ was as virulence determinants implicated in the ‘initiation of the systemic toxicity which linked with fierceness of diseases and severe infections caused by S. pyogenes, a smeZ gene was the most exhibiting an effective super antigen, contributed imperative role, and coded to highly mitogenic proteins produced by numerous isolates of S. pyogenes23. The recent data was difference from results of the study performed by24 documented that 79% of invasive isolates had this gene.
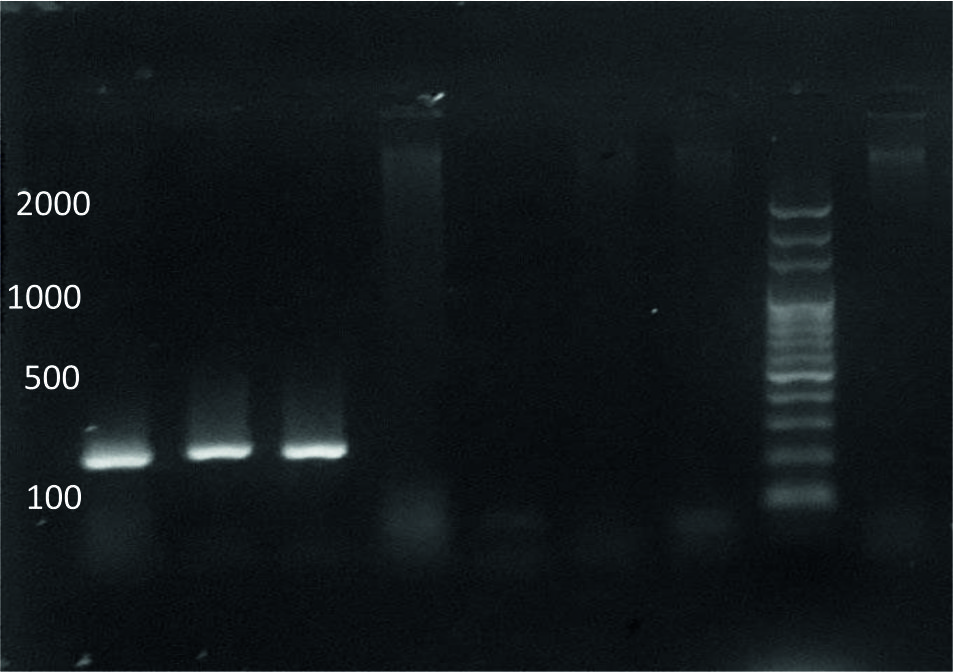
Fig. 2. Agarose gel electrophoresis of smeZgene amplification,M: ladder, 1-3: positive results, 4-7: negative results.
The changed distribution of smeZ gene may be associated with transferred this gene by different elements through chromosomal DNA. Furthermore, Schmitz et al.,25 showed the distribution rate of smeZ, speA, speC, speH, speJ and ssa genes were associated with movable elements, and the smeZ allele was found in 95.8% strains. The results of this study were a low emergence than the results of the study by26 recorded that all invasive strains harbored smeZ gene.
In the same field, the results of current data disagreed with the study of 27 documented a high emergence of isolates had this gene (99%).
‘The DNA sequences of specific gene (spy1258) was only presented in S. pyogenes and absent in other bacterial species’. Numerous studies had used the spy1258 for the fast detection of GAS that recovered from countless clinical samples’. Also the results of study performed by21 recorded that spy1258 gene was sensitive and specific for S. pyogenes, and this gene found in completely isolates of GAS (100%), while 19 revealed that the sensitivity of spy1258 gene by using of qPCR technique for direct detection of GAS isolated from throat19 . The study validated by28 Zhao et al., (2015) documented that the spy1258 gene used for accurate and precise identification of GAS strains,
Phylogenetic tree of S. pyogenes based on the neighbour-linking of partspy1258 and smeZ genes sequences presented those sequences were resulted from genes of Streptococcus (Fig 3 and 4).
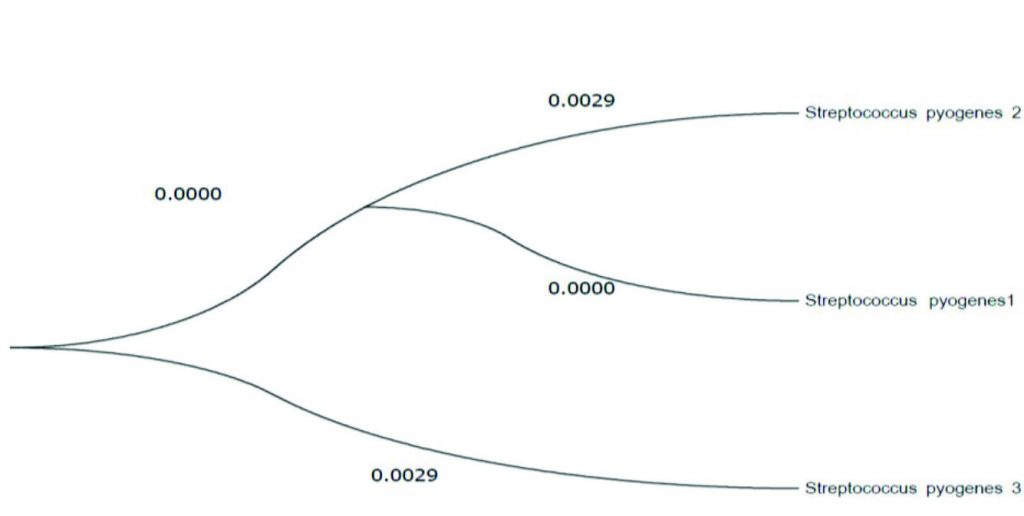
Fig. 3. The evolutionary relationships of S. pyogenes, phylogeny tree of the smeZ gene inferred through distance based analysis using Tamura-Nei distance estimates of aligned nucleotide sequences resulting from the PCR sequence data.
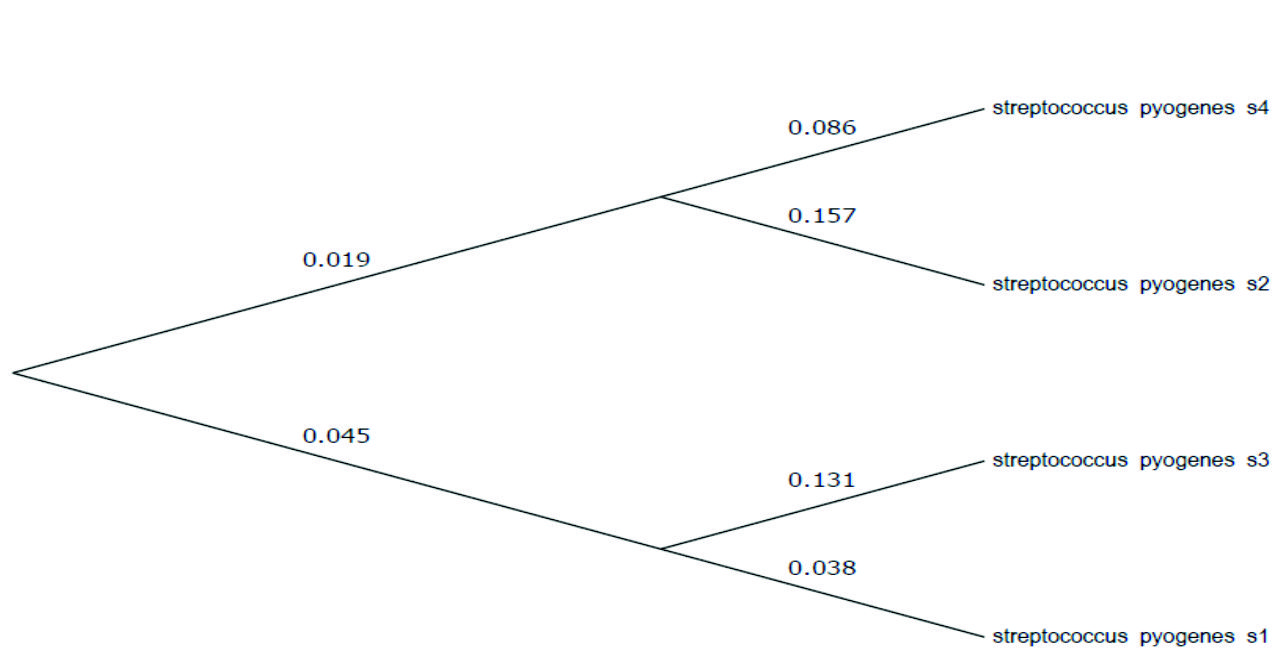
Fig. 4. The evolutionary relationships of S. pyogenes, phylogeny tree of the spy1258 gene was showedvia distance depended on analysis by Tamura-Nei distance estimates of allied nucleotide sequences consequent from the PCR sequence data.
To classify any organism such as bacteria used the phylogenetic analysis method29 showed the evolutionary history was inferred via the neighbor joining process. The best tree with the sum of branch-length = 0.47675007 was revealed (above the branches), while Maximum Composite Likelihood process was performed to compute the an evolutionary distances and in the units of the numeral of base substitutions of each site30 .
On the other hand, Babbar et al.,31 reported that the more examinations of phenotypic features and determination of bacterial species shed extra light on the microbial diversity like S. pyogenes and expanding the understanding the infections of this microbe, and its diagnosis.
The analysis including four nucleotide sequences. Codon locations involved were 1st + 2nd + 3rd + Noncoding. All vague positions were detached for each sequence pair, there were a entire of 220 locations in the ending dataset, and the evolutionary analyses were directed in MEGA732 .
The sequencing of PCR products produced for spy1258 gene showed significant alignments identities (100%) to Streptococcus pyogenes strain NCTC12696 genome assembly, chromosome: 1 Sequence ID: LS483332.1 which are located in BLAST-NCBI Genbank. While the alignments identities for smeZ gene was (94-100%) to S. pyogenes (ID: LS483384.1, LS483330.1 and LS483394.1) which are located in BLAST-NCBI Genbank.
From the results of Multiple sequence alignment analysis, showed C > T polymorphism in sample two of smeZ gene, Fig (5). While recorded T > C polymorphism in sample one, Fig (6). This variation in nitrogen base may be related for differentiation the product of current gene (smeZ) which may be related to increased pathogenicity of this microorganism that harbored in it.
The spy 1258 gene was identical gene for S. pyogenes, and used as useful tool for tonsillitis GAS rapid method to diagnosis.
The author would like to special thanks for Dr. Khwam R. Hussein, Sothern Technical University for their supports in submitted to DNA sequencing of studied genes in BLAST-NCBI Genbank.
None
- Rantala, S.; Va¨ha¨ kuopus, S.; Siljander, T.; Vuopio, J.; Huhtala, H.;Vuento, R. and Syrjanen, J. Streptococcus pyogenes bacteraemia, emm types and superantigen profiles. Eur. J. Clin. Microbiol. Infect. Dis., 2012; 31: 859–865.
- Hertze, E.; Johansson, L.;Wallin, R.; Schmidt, H.; Kroll, M.; Rehn, A P.; Kotb, M.; Mצrgelin, M. and Norrby-Teglund, A. M1 protein-dependent intra-cellular trafficking promotes persistence and replication of Streptococcus pyogenes in macro-phages. J. Inn. Immun., 2010; 2: 534–545.
- Staali, L.; Bauer, S.;Morgelin, M.;Bjorck, L. and Tapper, H. Streptococcus pyogenes bacteria modulate membrane traffic in human neutrophils and selectively inhibit azurophilic granule fusion with phagosomes. J. Cell. Microbiol., 2006; 8: 690–703.
- Walker, M J.;Hollands, A.; Sanderson-Smith, M L.; Cole, J N. ; Kirk, J K.; Henningham, A.; McArthur, J D.; Dinkla, K.; Aziz, R K.; Kansal, R G.; Simpson, A J.; Buchanan, J T.; Chhatwal, G S.; Kotb, M. and Nizet, V. DNase Sda1 provides selection pressure for a switch to invasive group A streptococcal infection. J. Nat. Med., 2007; 13: 981–985.
- Walker, M J.;Hollands, A.; Sanderson-Smith, M L.; Cole, J N. ; Kirk, J K.; Henningham, A.; McArthur, J D.; Dinkla, K.; Aziz, R K.; Kansal, R G.; Simpson, A J.; Buchanan, J T.; Chhatwal, G S.; Kotb, M. and Nizet, V. DNase Sda1 provides selection pressure for a switch to invasive group A streptococcal infection. J. Nat. Med., 2007; 13: 981–985.
- Walker, M J.; Hollands, A.; Sanderson-Smith, M L.; Cole, J N. ; Kirk, J K.; Henningham, A.; McArthur, J D.; Dinkla, K.; Aziz, R K.; Kansal, R G.; Simpson, A J.; Buchanan, J T.; Chhatwal, G S.; Kotb, M. and Nizet, V. DNase Sda1 provides selection pressure for a switch to invasive group A streptococcal infection. J. Nat. Med., 2007; 13: 981–985.
- Kumar, A.; Bhatnagar, A.; Gupta, A.; Khare, A. and Suman, A. Sofgene as a specific genetic marker for detection of Streptococcus pyogenes causing pharyngitis and rheumatic heart disease. J. Cell. Mol. Biol., 2011; 57: 26-30.
- Liu, D.; Hollingshead, S.; Swiatlo, E.; Lawrence, ML. and Austin, FW. Rapid identification of Streptococcus pyogenes with PCR primers from a putative transcriptional regulator gene. J. Res. Micro., 2005; 156: 564-567.
- Brahmadathan, KN. and Gladstone P. Micro-biological diagnosis of streptococcal pharyngitis: lacunae and their implications. India. J. Med. Micro., 2006; 24(2): 92-96.
- Harley, JP. and Prescott, LM. Laboratory Exercises in Microbiology. 5th ed. New York. McGraw-Hill, 2002.
- Abraham, T. and Sistla, S. Identification of Streptococcus pyogenes phenotypic tests vs molecular assay (spy1258PCR): A comparative study. J. Clin.Dia. Res.,2016; 10(7): 1-3.
- Borek, A L.; Obszaסska, K.; Hryniewicz, W. and Sitkiewicz, I. Detection of Streptococcus pyogenes virulence factors by multiplex PCR. J. Vir., 2012; 3(6):529–533.
- Hall, TA. Bioedit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98nt. Nucl Acids Symb ser., 1999; 41: 95-98.
- Kumar, S.; Stecher, G. and Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. J. Mol. Biol.Evol.; 2016; 33: 1870-1874.
- Kumar, S.; Stecher, G. and Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. J. Mol. Biol.Evol.; 2016; 33: 1870-1874.
- Beye, M.; El Karkouri, K.; Labas, N.; Raoult, D. and Fournier, P E. Genomic analysis of a Streptococcus pyogenes strain causing endocarditis in a child. J. NewMicrob. New Infect., 2017; 17:(1–6).
- Dakhil, B R. and Hamim, S S. Antibiotic susceptibility of Streptococcus pyogenes and Staphylococcus aureus isolated from pharyngitis and tonsillitis patients in Nasiriyah City, Iraq. World. J. Pharm. Sci., 2016; 4(4): 14-19.
- Abidali, ZN. (2014). Immunological and molecular study of the bacteria that cause Tonsillitis. Master thesis. College of Science for Girls/ Babylon University, Iraq.
- Dunne, E M.; Marshall, J L.; Baker, C A.; Manning, J.; Gonis, G.; Danchin, M H.;Smeesters, P R.; Satzke,C. and Steer, A C. Detection of group A streptococcal pharyngitis by quantitative PCR. J. Bio. Med. Cent. Infect. Dis., 2013; 13(312):1-7.
- Schabauer, L.; Wenning, M.; Huber, I.; and Ehling-Schulz, M. Novel physico-chemical diagnostic tools for high throughput identification of bovine mastitis associated gram-positive, catalase-negative cocci. J. BMC. veteran. Resea., 2014; 10(1):156.
- Al-Saadi, K A.; Naji, H F.; Al-Saadi, A H. and Ali, A H M. Detection and identification of Streptococcus pyogenes from ENT patients by different methods. J. Pharm. Biomed. Sci., 2015; 05(06): 480-486.
- Al-Shwany, Z H. and Al-Jebori, I S A. Molecular study of sortase enzyme in Streptococcus pyogenes isolated from patients with tonsillitis in Kirkuk city. J. Kirk. Uni.Sci. Stud. (KUJSS)., 2015; 10: 227-241.
- Norrby-Teglund, A.; Thulin, P.; Gan, B S.; Kotb, M.;McGeer, A.; Andersson, J. and Low, DE. Evidence for superantigen involvement in severe group A streptococcal tissue infections. J. Infect. Dis., 2001; 184: 853–860.
- Sriskandan, S.; Faulkner, L. and Hopkins, P. Streptococcus pyogenes: insight into the function of the streptococcal superantigens. J. Int.Biochem. Cell. Biol., 2007; 39: 12–19.
- Schmitz, F J.; Beyer, A.; Charpentier, E.; Normark, B H.; Schade, M.; Fluit, A C.; Hafner, D. and Novak, R. Toxin gene profile heterogeneity among endemicinvasive European group A Streptococcal isolates. J. Infect. Dis., 2003; 188: 1578–1586.
- Luca-Harari, B.; Straut, M.; Cretoiu, S.; Surdeanu, M.; Ungureanu, V.; van der Linden, M. and Jasir, A. Molecular characterization of invasive and noninvasive Streptococcus pyogenes isolates from Romania. J.Med. Micro., 2008; 57:1354–1363.
- Meisa, R.; Andreasson, I KG.; Hרiby, E A.; Aaberge, I S.; Michaelsen, T E. and Caugant, D A. Strep-tococcus pyogenes isolates causing severe infections in Norway in 2006 to 2007: emm Types, Multilocus sequence types, and superantigen Profiles. J. Clin.Microbiol., 2010; 48(3): 842–851.
- Meisa, R.; Andreasson, I KG.; Hרiby, E A.; Aaberge, I S.; Michaelsen, T E. and Caugant, D A. Strep-tococcus pyogenes isolates causing severe infections in Norway in 2006 to 2007: emmTypes, Multilocus sequence types, and superantigen Profiles. J. Clin.Microbiol., 2010 48(3): 842–851.
- Dewangan, R.; Patel, A.; Khatri, S.; Choubey, J.; Verma, M.K.; Kumar Gupta, S. and Rishi, V. Phylogenetic analysis of Enterococcus, Lacto-bacillus and Streptococcus strains on the basis of abc(Atp Binding Protein) gene sequences. Curr. Res. J. Biol. Sci., 2009; 1(3): 127-130.
- Dewangan, R.; Patel, A.; Khatri, S.; Choubey, J.; Verma, M.K.; Kumar Gupta, S. and Rishi, V. Phylogenetic analysis of Enterococcus, Lacto-bacillus and Streptococcus strains on the basis of abc(Atp Binding Protein) gene sequences. Curr. Res. J. Biol. Sci., 2009; 1(3): 127-130.
- Babbar, A.; Kumar, VN.; Bergmann, R.; Barrantes, I.; Pieper, DH.; Itzek, A. and Nitsche-Schmitz, DP. Members of a new subgroup of Streptococcus anginosus harbor virulence related genes previously observed in Streptococcus pyogenes. Int. J. Med. Microbiol., 2017; 307(3):174-181.
- Kumar, S.;Stecher, G. and Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. J. Mol. Biol.Evol.; 2016; 33: 1870-1874.
© The Author(s) 2019. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.