ISSN: 0973-7510
E-ISSN: 2581-690X
The most important products obtained for human needs are enzymes, through microbial sources. Hydrolytic enzymes occupied major industrial share such as esterases, lipases, amylases and proteases. Applications of these enzymes are varied in medical, cosmetics, food, dairy, pharmaceutical, leather, detergents, bioremediations and paper industries. So, lipases are gaining more attention now a day. Lipases hydrolyse fats into monoglycerides, diglycerides, free fatty acids and glycerol. Bacillus and Stenotrophomonas, are found to be the efficient lipase producers. A total of 48 different lipolytic organisms were isolated from soil samples. Organisms producing lipases were selected on the basis of clear zone formation in tributyrin agar medium. Maximum enzyme yield was obtained at 40°C with 16 U/ml in Bacillus and 14 U/ml in Stenotrophomonas sp., at a pH of 7 in 3 days. The optimum parameters were studied. The yield of lipase enzyme was high while using peptone as a nitrogen source in Stenotrophomonas sp., where as in presence of CaCl2, Bacillus sp., produced more lipase enzyme. However the presence of glucose as carbon source also yielded a much closer value to be highest yield. The lipase producing strains were subjected to gram staining and biochemical characterization following Bergey’s manual of systematic bacteriology. 16SrRNA studies and physiological characterization reveals that Bacillus sp., Stenotrophomonas sp., Halomonas sp., Serratia sp., and Enterobacter sp., are the efficient producers. The sequences were deposited in Genbank.
Enzymes, Lipase, Bacillus, Stenotrophomonas, Optimization, Applications.
Lipase produced from bacteria is an important enzyme in food, detergents and pharmaceutical industries [Abu et al., 2005]. These are very important in environmental managements [Aravindan et al., 2007]. All other organisms including humans, animals, and plants depend upon them, without microorganisms no life can exist on earth [Bajpai and Tyagi, 2007]. Microorganisms are capable of tolerating wide range of conditions, so they are great interest as small biofactories. Hydrolases include lipases, proteases, pectinases and carbohydrases [Fujii et al., 1986]. Lipases are fat splitting enzymes which require excess water to catalyze the formation of glycerol and fatty acids from long-chain triacylglycerol’s by hydrolysis [Jaeger et al., 1994, Pallavi et al., 2010 and Ram Reddy & Pallavi, 2012]. Lipases produced by bacteria are used in dairy industries to hydrolyze milk fat, cheese ripening and flavor enhancement. Lipases are also used as additives in detergent industry. [Basketter et al., 2008, Falch, 1991]. Mostly microbial lipases are extracellular and production is influenced by media composition with various physico chemical factors such as pH, temperature and dissolved oxygen. [Gulati et al., 2005, Gilman, 1998]. Optimization of culture medium achieved high productivity. [Podar and Reysenbach, 2006].
Collection of Sample
Samples were collected from oil contaminated soils around Salem, Coimbatore, Vellore and Chennai. Collected Samples were taken to the laboratory and refrigerated at 4°C.
Isolation and Identification of Bacteria producing lipase
Soil samples were serially diluted and 0.1ml from 10-4 and 10-5 dilutions were plated on nutrient agar. Identification of organisms was done based on morphological and biochemical characters following Bergey’s manual of Systemic Bacteriology [Sneath, 1986]. Organism producing lipase were identified by formation of clear zone around the colony in tributyrin agar, after 48 – 72 hrs of incubation at 37°C. Clear zone producing colonies were picked and stored on nutrient agar slants for further studies. [Mobarak – Qamsaari et al., 2011].
Submerged Fermentation
Medium for production of enzyme consists of peptone 0.2, NH4H2PO4 0.1, NaCl2 0.25, Olive oil 2 (V/V), Cacl22H2O 0.04, MgSo4 7H2O 0.04, pH 7, 1-2 drops of Tween 80 as emulsifier for 100ml distilled water. 100ml of inoculated media were kept in rotary shaker at 150 rpm for 24hrs. After inoculation, the broth culture was centrifuged at 10,000 rpm for 10min and placed at 4°C. The cell fitrate is the source of extracellular enzyme. [Aliyu et al., 2011].
Enzyme Assay
Lipase activity was demonstrated by using spectrophotometrically at 30°C by using p-nitrophenol palmitate (pNPP) as a substrate [Winkler and Stuckmann, 1979]. Reaction mixture contains 300μl of lipase solution with 700μl pNPP solution. Preparation of pNPP by addition of solution A (0.001 g pNPP in 1ml isopropanal) in solution B (0.01 g gum arabic, 0.02 g Sodium deoxycholate, 50μl Triton X-100 and 9 ml of 50 mM Tris-HCl buffer, pH 8). Measured the absorbance at 410 nm by first 2 min of reaction. One unit was defined as the amount of enzyme liberated 1μmol of pNP per minute under laboratory conditions [Karadzic et al., 2006, Rathi et al., 2001, Tyagi et al., 2002, Lubna et al., 2015].
Optimization of lipase production
Production of lipase was carried out at different pH (4-8), temperature (30°- 70°C) and incubation period (3-7 days) [Kumar et al., 2012]. Various metal ions such as calcium chloride, sodium chloride, magnesium sulphate, ferrous sulphate and zinc chloride, various carbon sources such as maltose, mannitol, lactose, sucrose and glucose, Nitrogen sources such as yeast extract, peptone, casein, albumin and urea are separately added to a final concentration of 1% (w/v) to media and fermented. After incubation enzyme activity was observed as described above. [Gupta et al., 2004, Kim et al., 1996, Gao et al., 2000, Chen et al., 1998].
Oil Source and Surfactants
Production of lipase was enriched by addition of various lipid sources such as coconut oil, groundnut oil, olive oil, castor oil and neem oil and surfactants such as Tween 20, Tween 40, Tween 60, Tween 80, Tween 100, Triton X – 100, SDS) were added separately and tested at a concentration of 1% in production medium.
Identification and Molecular characterization of the isolates
The lipase producing strains were subjected to gram staining and biochemical characterization using standard methods. 16S rRNA was done to confirm the organisms using 27F and 1492R. Studies and physiological characterization reveals that Bacillus sp., Stenotrophomonas sp., Halomonas sp., Serratia sp., and Enterobacter sp., are the efficient producers. The sequences were deposited in Genbank.
Isolation and Identification of Lipolytic Microorganisms
Forty eight microorganisms were isolated from soil samples based on the formation of clear zones on tributyrin agar. (Fig. 1). Sixteen organisms were selected for further study selected based on wider zones on tributyrin agar. The results were given in the Table no.1.
Table (1):
Biochemical characterization of the isolates.
S.No |
Strains |
Gram staining |
Motility test |
Indole test |
MR test |
VP test |
Citrate test |
Catalase test |
Oxidase test |
Urease test |
Carbohydrate Fermentation test |
|---|---|---|---|---|---|---|---|---|---|---|---|
1 |
Bacillus sp., |
+ |
+ |
_ |
_ |
+ |
+ |
+ |
_ |
_ |
+ |
2 |
Pseudomonas sp., |
– |
+ |
– |
_ |
_ |
+ |
+ |
+ |
_ |
_ |
3 |
Staphylococcus sp., |
+ |
_ |
_ |
+ |
+ |
+ |
+ |
_ |
+ |
+ |
4 |
Streptomyces sp., |
+ |
– |
– |
– |
+ |
+ |
– |
+ |
+ |
_ |
5 |
Klebsiella sp., |
– |
– |
– |
– |
+ |
+ |
+ |
– |
+ |
+ |
6 |
Proteus sp., |
– |
+ |
– |
+ |
_ |
+ |
+ |
– |
+ |
_ |
7 |
Serratia sp., |
– |
+ |
– |
– |
+ |
+ |
+ |
– |
+ |
_ |
8 |
Lactobacillus sp., |
+ |
– |
– |
+ |
– |
– |
– |
+ |
– |
+ |
9 |
Streptococcus sp., |
+ |
– |
– |
– |
– |
– |
– |
– |
– |
+ |
10 |
Aeromonas sp., |
– |
+ |
+ |
– |
+ |
+ |
+ |
+ |
– |
_ |
11 |
Micrococcus sp., |
+ |
– |
– |
– |
– |
– |
+ |
+ |
+ |
_ |
12 |
Arthrobacter sp., |
+ |
– |
– |
– |
– |
– |
+ |
– |
— |
_ |
13 |
Escherichia coli |
– |
+ |
+ |
+ |
– |
– |
+ |
– |
– |
+ |
14 |
Halomonas sp., |
– |
+ |
+ |
– |
+ |
– |
+ |
+ |
– |
+ |
15 |
Enterobacter sp., |
– |
+ |
– |
– |
+ |
+ |
+ |
– |
– |
_ |
16 |
Stenotrophomonas sp., |
– |
+ |
– |
– |
– |
+ |
+ |
– |
– |
_ |
Enzyme assay
Among 48 organisms, the selected 16 organisms were subjected to production of lipase. The enzyme activities of all the isolates are shown in table 2. Among 16 isolates, 5 isolates (Bacillus sp., Halomonas sp., Enterobacter sp., Serratia sp., Stenotrophomonas sp.,) were able to produce lipase above 11U/ml when incubated for 72 Hrs at pH 7 at 37°C. On the basis of production rate, out of 16 organisms, 5 isolates were selected for further studies.
Table (2):
Lipase enzyme production in submerged fermentation (SSF) by selected Bacterial species.
S.No |
Strains |
Lipase Activity(U/ml) SSF |
|---|---|---|
1. |
Bacillus sp., |
14.03±0.75 |
2. |
Staphylococcus sp., |
9.03 ±0.850 |
3. |
Escherichia coli |
4.86 ±0.808 |
4. |
Micrococcus sp., |
7.96 ± 0.85 |
5. |
Streptomyces sp., |
07 ±1 |
6. |
Klebsiella sp., |
3.93 ±0.901 |
7. |
Serratia sp., |
13 ±1 |
8. |
Proteus sp., |
10 ±1 |
9. |
Pseudomonas sp., |
9.833±1.25 |
10. |
Enterobacter sp., |
12 ±1 |
11. |
Aeromonas sp., |
07 ±1 |
12. |
Pseudomonas sp., |
10.83 ±0.763 |
13. |
Micrococcus sp., |
05 ±1 |
14. |
Halomonas sp., |
12 ±1 |
15. |
Stenotrophomonas sp., |
14.83 ±0.763 |
16. |
Streptococcus sp., |
9.01 ±0.6 |
Optimization of enzyme production
Effect of temperatures and pH on enzyme production
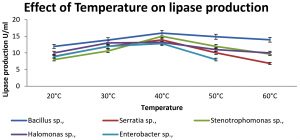
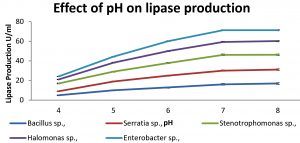
Production of lipase at different temperatures (20 – 60°C) was observed for 72 hrs. Lipase activity was maximum at 40°C in all the isolates. The enzyme activities at different temperatures are shown in Fig. 2. Effect of pH on enzyme activity was shown in Fig. 3. Maximum production of lipase was obtained at pH 7 for Serratia sp., and. Stenotrophomonas sp., whereas for Bacillus and Halomonas sp., at pH 8.
Effect of Incubation Periods
Production of lipase by isolated organisms after incubation at different time periods ranging from 12 – 72 hrs were assayed. Bacillus sp., shows maximum production of enzyme at 24 hrs with 18U/ml. Serratia sp., produce 14U/ml of enzyme at 24 – 48Hrs. Stenotrophomonas sp., produce 17U/ml at 24hrs. Halomonas sp., produce maximum lipase activity at 24hrs with 14U/ml. Enterobacter sp., produce maximum activity of 14U/ml at 24 hrs. Table no: 3 Maximum [Mahanta et al., 2008] production of lipase by Bacillus stearothermophilus occurred at 24hr of incubation. Decrease in production of enzyme may be due to interaction of enzyme with other compounds in medium or due to reduction of nutrients in batch cultures.
Table (3):
Effect of Incubation periods on lipase production.
| Incubation periods | Bacillus sp.,(U/ml) | Serratia sp.,(U/ml) | Stenotrophomonas sp.,(U/ml) | Halomonas sp.,(U/ml) | Enterobacter sp., U/ml) | |
|---|---|---|---|---|---|---|
| 12hrs | 13.06±0.3 | 9.3±0.9 | 8.16±0.5 | 10.16±0.5 | 9.1±0.65 | |
| 18hrs | 14.93±0.7 | 11.9±0.4 | 13.93±0.4 | 12.83±0.4 | 11.96±0.45 | |
| 24hrs | 17.96±0.7 | 13.8±0.7 | 17.1±0.6 | 13.96±0.5 | 13.93±0.7 | |
| 48hrs | 14.86±0.8 | 14.2±0.5 | 14.9±0.4 | 13±0.5 | 13.06±0.5 | |
| 60hrs | 10.2±0.5 | 9±0.7 | 8.2±0.6 | 11.96±0.4 | 8.93±0.7 | |
Effect of various carbon, nitrogen sources and metal ions
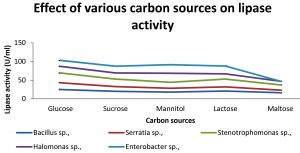
Various carbon sources such as glucose, sucrose, mannitol, lactose and maltose are used in media for production of enzyme. The effect of different carbon sources are shown in Fig 4. Glucose as a carbon sources was found to be the optimum for Bacillus sp., Serratia sp., and Stenotrophomonas sp.,while mannitol was found to be the best carbon sources for Halomonas sp.,and Enterobacter. Ghaima et al., (2014) had reported that with maltose as the carbon source, B. cereus had the highest activity however in our study high yield was obtained with glucose in B. cereus.
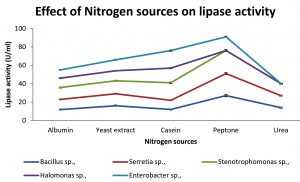
Various nitrogen sources such as peptone, yeast extract, casein, Albumin and urea are assayed for maximum production of lipase. The effect of nitrogen sources are shown in Fig: 5. Peptone is known as common inducer for lipase. The releasing of NH4+ ions from peptone influenced its efficacy for higher enzyme activity because NH4 stimulates the growth and increases enzyme production rate [Mukesh Kumar et al., 2012]. Peptone was found to be best nitrogen source for the isolates except Enterobacter sp., which produced maximum lipase with casein.
Various metal ions such as MgSo4, CaCl2, FeSo4, NaCl2 and ZnCl2 are assayed for production of lipase.CaCl2 enhanced the production of lipase by the organisms. The effects of different ions are shown in table. 4. Extracellular lipase produced by Bacillus sp., yield high activity of lipase in presence of Ca2+ which is in agreement with several previous reports. [Jayaraman, 1981, Bajpai and Tyagi, 2007].
Table (4):
Effect of Metal ions on lipase production.
| Metal ions | Bacillus sp.,(U/ml) | Serratia sp.,(U/ml) | Stenotrophomonas sp.,(U/ml) | Halomonas sp.,(U/ml) | Enterobacter sp.,U/ml) | |
|---|---|---|---|---|---|---|
| MgSo4 | 24.9±0.36 | 19.9±0.45 | 20.1±0.36 | 18.03±0.35 | 18.9±0.36 | |
| CaCl2 | 27±0.5 | 19.1±0.61 | 28.1±0.55 | 19.9±0.4 | 22.13±0.15 | |
| FeSo4 | 17.96±0.35 | 13±0.5 | 18.03±0.45 | 13±0.5 | 17.9±0.4 | |
| NaCl2 | 16.06±0.6 | 15±0.5 | 19±0.5 | 16±0.5 | 19.8±0.32 | |
| ZnCl2 | 16±0.5 | 9.9±0.5 | 16.03±0.5 | 12±0.5 | 20.03±0.25 | |
Influence of oils and surfactants
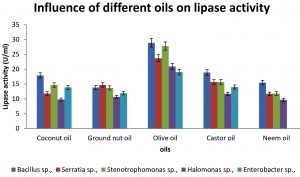
Lipases are induced mostly in presence of oils in culture medium. Maximum lipase production was seen in presence of olive oil (29U/ml) in Bacillus sp., Stenotrophomonas sp., (28U/ml), Enterobacter sp., (19U/ml) Halomonas sp., (21U/ml) and Serratia sp., (24U/ml). Similar studies conducted on lipase production by Bacillus sp., showed maximum yield in olive oil 8. Various studies revealed that olive oil is more effective to induce production of lipase, due to lipid content in olive oil, which is structurally easily digested by microorganisms [Zarevucka, 2012].
Table (5):
Influence of surfactants on lipase production.
| Surfectants | Bacillus sp.,(U/ml) | Serratia sp.,(U/ml) | Stenotrophomonas sp.,(U/ml) | Halomonas sp.,(U/ml) | Enterobacter sp.,U/ml) | |
|---|---|---|---|---|---|---|
| Tween 20 | 14.03±0.45 | 8.16±0.37 | 13.03±0.15 | 10±0.3 | 9.03±0.35 | |
| Tween 40 | 10.2±0.52 | 11.93±0.4 | 10±0.2 | 8.1±0.5 | 9.9±0.35 | |
| Tween 60 | 19.16±0.56 | 16±0.5 | 19.06±0.3 | 13.06±0.3 | 10±0.2 | |
| Tween 80 | 30±0.2 | 23.03±0.45 | 25±0.5 | 19.06±0.4 | 20.06±0.3 | |
| Tween 100 | 21.13±0.3 | 20.06±0.3 | 18.1±0.55 | 14.03±0.45 | 14.03±0.45 | |
| Triton X-100 | 16±0.5 | 16.06±0.5 | 10.1±0.45 | 13.03±0.45 | 16.03±0.45 | |
| SDS | 12.06±0.3 | 14.9±0.3 | 11.03±0.15 | 11.9±0.35 | 13±0.4 | |
Fermentation medium enhance the secretion of proteins by alterating the cell membrane permeability, so addition of surfactants enhances production of lipase. Tween-80 showed maximum yield of lipase in Bacillus sp., (30 U/ml) Stenotrophomonas sp., (25 U/ml) Halomonas sp., (19 U/ml) Serratia sp., (23 U/ml) and in Enterobacter sp., (20 U/ml). The effect of surfactants on lipase production is shown in fig.6. Thus our results in accordance with Duttat and Ray, 2009 who had shown that surfactants have no or little inhibitory activity on enzyme activity.
Molecular characterization of the isolates
Based on 16S rRNA sequencing the five isolated organisms are identified as Bacillus cereus, Serratia marcescens, Stenotrophomonas maltophila, Serratia marcescens and Enterobacter cloaca. The accession numbers of the sequences deposited in NCBI are MNO94422, MK999992, MNO94371, MNO94423 and MK999988 respectively.
Lipase is one of the most valuable industrial enzymes. In present study, lipase producing organisms are isolated from soil sample. 48 organisms was isolated from soil sample and optimized with various physico – chemical parameters for lipase production. The enzyme produced by B. cereus was found to be stable at 60°C though its optimum is 40°C. A novel strain Stenotrophomonas maltophila was found to produce lipase with the simple sugar glucose. Also it is observed that these enzymes remain active at alkaline pH. Because of this, these enzymes could be promising biocatalyzing agents in industries.
ACKNOWLEDGMENTS
The authors are thankful to the Management and the Principal, Hindusthan college of Arts and science college, Coimbatore, Tamil Nadu, India, for providing us the laboratory facilities for research.
CONFLICT OF INTEREST
The authors declare that there are no conflicts of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work and approved it for publication. Joyruth performed the experiments and prepared the manuscript. LG provided technical guidance and reviewed the manuscript.
FUNDING
None.
ETHICS STATEMENT
The research does not involve human or animal experimentation.
AVAILABILITY OF DATA
All available data studied are incorporated in the manuscript.
- Abu EA, Ado SA and James DB. Raw starch degrading amylase production by mixed cultures of Aspergillus niger and Saccharomyces cerevisae grown on sorghum pomace, African J. Biotechnol., 2005; 4: 785 – 790.
- Aliyu AB et al, Activity of saponin fraction of Anisopus mannii against some pathogenic microorganisms. Journal of Medicinal Plant Research, 2011; 5(31): 6709 – 6713.
Crossref - Aravindan R, Anbumathi P and Viruthagiri T. Lipase applications in food industry. Indian Journal of Biotechnology, 2007; 6: 141 – 158.
- Bajpai D and Tyagi VK. Laundry detergents: an overview, 2007; 56: 327 – 340.
Crossref - Basketter DA, English JS, Wakelin SH and White IR. Enzymes, detergents and skin. Facts and fantasies. Br. J. Dermatol., 2008; 158: 1177-1181.
Crossref - Chen SJ, Cheng CY and Chen TL. Production of an alkaline lipase by Acinetobacter radioresistens. J.Ferment Bioeng., 1998; 86(3): 308 – 312.
Crossref - Dutta S and Ray L. Production and characterization of an alkaline thermostable crude lipase from an isolated strain of Bacillus cereus C7. Appl. Biochem. Biotechno., 2009; 159: 142 – 154.
Crossref - Ewis HE, Abdelal AT and Lu CD. Molecular cloning and characterization of two thermostable carboxyl esterases from Geobacillus stearothermophilus.
Gene, 2004; 329: 187-195.
Crossref - Falch EA. Industrial enzymes – developments in production and application. Biotechnol. Adv., 1991; 9(4): 643 – 658.
Crossref - Fujii T, Tatara T and Minagawa M. Studies on application of lipolytic enzyme in detergent industries. Effect of lipase from Candida cylindracea on removal of olive oil from cotton fabric. J. Am. Oil Chem. Soc., 1986; 63: 796 – 799.
Crossref - Gao XG, Cao SG and Zhang KC. Production, properties and application to non aqueous enzymatic catalysis of lipase from a newly isolated Pseudomonas strain. Enzyme Microb Technol., 2000; 27: 74 – 82.
Crossref - Ghaima KK, Mohamed AI and Mohamed MM. Effect of some factors on lipase production by Bacillus cereus isolated from diesel fuel polluted soil. International Journal of scientific Research publication, 2014; 4(8): 188 – 202.
- Gilman CJ. A manual of soil fungi. Biotech books. New Delhi, 1998.
- Gulati R et al. Production of a novel alkaline lipase by Fusarium globulosum using neem oil and its applications. Pure App., Chem., 2005; 77(1): 251-262.
Crossref - Gupta R, Gupta N and Rathi P. Bacterial lipases: aqn overview of production, purification and biochemical properties. Appl. Microbial. Biotechnol., 2004; 64: 763 – 781.
Crossref - Hasan F, Shah AA and Hameed A. Industrial Applications of microbial lipases. Enzyme Microd. Technol., 2006; 39: 235 – 251.
Crossref - Jaeger KEH et al, Bacterial lipases. FEMS Microbial. Rev., 1994; 15: 29 – 63.
Crossref - Jayaraman J. Laboratory manual in Biochemistry. Wiley Eastern limited, New Delhi, 1981; 133.
- Joyruth P and Lali Growther. A Review on microbial lipase production, purification, characterization and its applications in various fields. Asian J. of Microbiol. Biotech. Env. Sc., 2018; 20(1): 294 – 300.
- Kambourova M, Kirilova N, Mandeva R and Derekova A., Purification and properties of thermostable lipase from a thermophiic Bacillus stearothermophilic MC. J. Mol. Catal. B. Enzyme., 2003; 7(22): 307-313.
Crossref - Karadzic I, Masui A, Lidija IZLI and Fujiwara N. Purification and characterization of an alkaline lipase from Pseudomonas aeruginosa isolated from putrid mineral cutting oil as component of metal working fluid. J. Biosci., Bioeng., 2006 ; 102: 82 – 89.
Crossref - Kim SS, Kim EK and Rhee JS. Effects of growth rate on the production of Pseudomonas fluorescens lipase during the fed – batch cultivation of Escherichia coli. Biotechnol. Prog., 1996; 12: 718-722.
Crossref - Kumar A, Sharma P & Kanwar SS. Lipase catalyzed esters syntheses in organic media: A Review. Int. J. Inst. Pharm. Life Sci., 2012; 2(2): 91 – 119.
- Lubna SA, Wells H and Fakhr MK. Staphylococcus aureus is more prevalent in retail beef livers than in pork and other beef cuts. Pathogens, 2015; 4: 182 – 198.
Crossref - Mahadik ND, Puntambekar US, Bastawde KB, Khive JM and Gokhale DV. Production of acidic lipase by Aspergillus niger in SSF. Process Biochem., 2002; 38: 715-721.
Crossref - Mahanta N, Gupta A and Khare SK. Production of protease and lipase by solvent tolerant Pseudomonas aureginosa PseA in solid – state fermentation using Jatropha curcas seed as substrate. Bioresour. Technol. 2008; 99: 1729 – 1735.
Crossref - Mobarak – Qamsaari E, Kasra Kermanshahi R and Moosavi – nejad Z, Isolation and identification of a novel, lipase – producing bacterium Pseudomonas aeruginosa KM110. Iran Journal of Microbiology, 2011; 3: 92 – 98.
- Mohan T, Palavesam A and Immanvel G. Isolation and characterization of lipase producing Bacillus strains from oil mills waste. African Journal of Biotechnology. 2008; 7: 2728-2735.
- Mukesh Kumar DJ et al, Characterization of lipase and protease from Serratia marcescens DEPTK21 and its destaining capability. Asian J. Exp. Biol. Sci., 2012; 3(3): 621-628.
- Pallavi P, Suresh A, Srinivas P and Ramreddy P. Optimization lipase production by staphylococcus sp., LP12. African J. Biotechnol., 2010; 9(6): 882 – 886.
Crossref - Podar M and Reysenbach A. New opportunities revealed by biotechnological explorations of extremophiles curr. Opin. Biotechnol., 2006; 17: 250-255.
Crossref - Ram Reddy S and Pallavi P. Microbial lipases – An overview In: Microbial diversity: Exploration and bioprospecting, Ram Reddy S, Singara charya MA and Girisham S(Eds.), Scientific Publishers (India), Jodhpur, 2012; 126 – 153.
- Rathi P, Saxena RK and Gupta R. A novel alkaline lipase from Burkholderia cepacia for detergent formulation. Process Biochemistry, 2001; 37: 187 – 192.
Crossref - Rosenau F and Jaeger KE. Bacterial lipases from Pseudomonas: Regulation of gene expression and mechanisms of secretion. Biochemie., 2000; 82: 1023-1032.
Crossref - Sharma SK, Olen CE and Parmar VS. Lipase catalyzed synthesis of optically enriched a – haloamides. Bioorg. Med. Chem., 2001; 9: 1345 – 1348.
Crossref - Sneath PHA. Bergeys Manual of Determinative Bacteriology, Williams and Wilkins, 1986; 2: 1105 – 1138.
- Treichel H, D de oliveira MA, Mazutti M, di Luccio and oliveira JV. A review on microbial lipases production. Food bioprocess Technology, 2010; 3: 182 – 196.
Crossref - Tyagi RD et al,Simultaneous production of Biopesticides and alkaline protease by Bacillus thuringiensis using sewage sludge as raw material. Water. Sci. Technol., 2002; 46: 247 – 254.
Crossref - Winkler and Stuckmann, Glycogen, hyaluronate and some other polysaccharides greatly enhance the formation of exolipase by serratia marcescens. J. Bacteriol., 1979; 138: 663 – 670.
Crossref - Zarevucka M. Olive oil as inductor of microbial lipase (Institute of organic chemistry and Biochemistry: Intech Europe). Olive oil improved the production of membrane bound lipase, 2012.
Crossref
© The Author(s) 2020. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.