ISSN: 0973-7510
E-ISSN: 2581-690X
Bioluminescent bacteria (BLB) are the most widely distributed light-emitting microorganisms, most of which are found as symbionts of free-living marine organisms, including the crustacean species. In this study, 4 out of 7 marine crustaceans in the northeastern section of Manila Bay were found to contain bioluminescent bacteria. Marine crustacean species namely Thenus orientalis (Oriental Slipper Lobster), Oratosquilla oratoria (Mantis Shrimp), Penaeus monodon (Giant Tiger Prawn), Litopenaeus vannamei (Whiteleg shrimp), Scylla serrata (Mud Crab), Portunus pelagicus (Blue Swimming Crab), and Charybdis feriata (Crucifix crab), were swabbed after collection. Bioluminescent bacteria were isolated from the inner surface of the carapace, eyes, abdomen, and abdominal segments of the crustacean samples. All glowing colonies were isolated and upscaled. Only bioluminescent bacteria from T. orientalis, O. oratoria, and P. monodon completed the isolation process and were identified using 16S rRNA gene sequencing for phylogenetic analysis. Furthermore, results from molecular identification through Nucleotide BLAST identified that it was Vibrio alginolyticus and Mucus bacterium. At the same time, the presence of Vibrio rumoiensis was also identified which was proved to be understudied and needs exploration. This study was aimed to identify the present bioluminescent bacteria in selected marine crustaceans in Manila Bay, Philippines through microbiological isolation, molecular identification, and phylogenetic reconstruction.
Vibrio alginolyticus, Vibrio rumoiensis, Marine Crustaceans, Manila Bay, 16S rRNA Gene Phylogeny
Vibrionaceae is the most abundant family of bacteria which includes several species of microorganisms that cause pathogenic intestinal tract infection following consumption of contaminated food and water by both humans and animals.1 Vibrionaceae symbiosis is widespread whether it is mutualism, host-pathogen relationship, or commensalism. They are also a part of marine sediments and form a sand biofilm microflora, and are attached to debris, zooplankton, phytoplankton, and carrion.2 V. harveyii is the most common causative agent of vibriosis affecting the fishes and crustaceans. Other Vibrio strains including V. vulni cus, V. alginolyticus, V. uvialis, V. mimicus, and V. parahaemolyticus are present in most marine crustacean species.3 In aquaculture, pathogenic strains of Vibrio sp. cause mortality of Penaeus monodon in hatcheries and coastal areas, which initiates a wide economic loss in agriculture and danger to the public health. V. alginolyticus, V. harveyii, V. anguillarium, and V. parahaemolyticus are the major causing species.4,5 Numerous cases of economic loss and risk to public health were documented caused by Vibriosis outbreaks on both human and animal. Consumption of raw or undercooked seafood and water which are exposed to the natural environment are highly associated with diarrhea, and other intestinal infections.6 Bioluminescent Vibrio sp. is one of the major etiological agents associated with the vibriosis in the pond-cultured P. monodon and its pathogenicity was obtained through the intramuscular injection to shrimp. The most dominant luminescent Vibrio species from P. monodon was V. harveyii, followed by V. logei, Photobacterium sp., and V. orientalis.7
Some of the Vibrio spp. in the family Vibrionaceae could produce a bluish-green cold light known as bioluminescence. These bioluminescent organisms produce light through a chemical reaction with the help of an enzyme and proteins in the presence of oxygen.8 These bacteria, either wild-type or genetically engineered bacteria, produce and emit cold light for communication, capturing prey, attracting mates and for predatory activities.8-11 Bioluminescent bacteria may be present in some terrestrial and aquatic organism such as annelids and worms, fungi, dinoflagellates, cnidarians, crustaceans, tunicates, echinoderms, and mollusks, which is regulated by different biochemical reactions.10-14 These bacteria act as gut symbionts in the digestive tracts of marine fishes, and some act as free-living in marine and brackish water ecosystems.15-17 They modified their forms and functions to adapt to the challenging environment; for example, the bioluminescence of some species is used as their cloaks of invisibility for security and some were used for communication.16,14
Bioluminescent bacteria could be used in different applications, they could be used as the toxin detection in water due to their high sensitivity to pollution,18,19 and they could also be used for analytical work in bioluminescent imaging and cancer cell therapies, and other biomedical applications for the detection of infection, and malignancies or cancers, because of their enzymes (luciferase) and reactants which are induced to the bacteria to produce a light signal.15,20-23 Recent findings from different studies done in the past showed that extracted secondary metabolites from Vibrio spp. contain antibiotic properties against gram-positive and gram-negative nosocomial bacteria.24 This report can be explored to alleviate the current threats of antibiotic resistance and nosocomial infections in clinical settings in the Philippines.
Food and waterborne diseases are the most common outbreaks which remain the leading causes of morbidity and mortality to the public health of the Philippines. In unclean water, microorganisms, toxic substances, and heavy metals mostly cause it. For an instance, in the Philippine Island of Western Mindanao, the villagers have the common symptoms including memory loss, heart and respiratory complications which are signs caused by mercury poisoning of “Minamata Disease”. This has been caused by the waste from abandoned mining area that was distributed among the landfill and water sources.25 Luminescent Vibrio sp. has high sensitivity to different concentration of heavy metals which are normally found in the contaminated water and soil. These bacteria could be utilized as biosensor for examining the quality of water, detection of heavy metals and toxic substances present in soil and water.18,19
The main objective of the study was to isolate and identify the bioluminescent bacteria that can be found in selected marine crustaceans from Manila Bay, Philippines based on molecular data and phylogenetic relationships. The study may contribute to provide more data on the presence of pathogenic bioluminescent bacteria in marine aquatic organisms. Moreso, this study may contribute to greater knowledge of the pond-culture fishermen on early detection and prevention of these disease-causing pathogens to their fishes and crustaceans that may lead to economic loss.
Sample collection of marine crustacean
The marine crustaceans were collected from Navotas Fishport Complex, Navotas City (14°38’51.797″ N, 120°56’52.331″ E) northeastern section of Manila Bay. It is the first fishing port that was placed under supervision of the Philippine Fisheries Development Authority and is dubbed as the fishing capital of the Philippines.
Morphological identification of the marine crustaceans
Each freshly caught marine crustacean species was identified based on the standard protocol for morphological identification of the fish samples using taxonomic keys and morphometric analysis. The verification was based on the journal website of World Register of Introduced Marine Species (WRiMS) and Global Biodiversity Information Facility (GBIF) for the appropriate classification of each crustacean sample.26
Isolation and purification of bioluminescent bacterial isolates
Seven crustacean samples: O. oratoria (Mantis shrimp), T. orientalis (Slipper lobster), P. monodon (Giant tiger prawn), S. serrata (Mud crab), P. pelagicus (Blue swimming crab), C. feriata (Crucifix crab), and P. vannamei (Whiteleg shrimp) were dissected and specific parts -the inner surface of carapace, eyes, abdomen, abdominal segments were swabbed separately. The cotton swabs were streaked on standard formulation of bioluminescent agar components: NaCl, peptone, K2HPO4, MgSO4, glycerol, agar and distilled water,19,24 incubated at 28oC for 24 hours. Bioluminescent colonies were picked and purified by standard microbiological streaking.
Bacterial genomic DNA extraction
A 1 mL of fresh upscaled bioluminescent bacterial cells with estimated cell density (12.0 x 108) based on McFarland turbidity standards were collected. The instructions of the QIAPrep® Spin Miniprep Kit were used to extract the genomic DNA (gDNA) of the collected bioluminescent bacterial cells. A 3µL isolated gDNA from the samples was run on a 1.2% agarose gel in 100V for 40 mins with DNA marker 1kb plus ladder (Invitrogen).
16S rRNA gene amplification
Gene amplification by PCR, components include genomic DNA, 27F (5’-AGAGTTTGATCCTGGCTCAG-3’) and 1492R (5’GGTTACCTTGTTACGACTT-3’) universal 16S primers.27 Cycling parameters on thermal cycler: 95°C 5 min; 30 cycles of 95°C 1 min, 60°C 45 secs, 70°C 1 min; 72°C 10 min; hold at 4°C. Post-PCR amplicon exhibited faint smear. Samples were purified prior to sequencing.
Sequencing and data analysis
Sanger di-deoxy sequencing was carried out. Cycle sequencing involves the incorporation of fluorescently labelled chain terminator ddNTPs; components include: amplicons, primers, and ABI BigDye® Terminator v3.1 Cycle Sequencing Kit. The cycling parameters on Bio-Rad T100 Thermal Cycler: pre-hold at 4°C; 96°C in 1 minute; 25 cycles of 96°C in 10 seconds, 50°C in 5 seconds, 62°C in 4 minutes; hold at 4°C. Ethanol precipitation was added to remove unincorporated ddNTPs, excess primers and primer dimers. Capillary electrophoresis was done on the ABI 3730xl DNA Analyzer using a 50cm 96-capillary array, POP7TM Polymer, and 3730xl Data Collection Software v3.1base calling on the Sequencing Analysis Software v5.4.
Quality control was done by initially converting the raw AB1 files to FASTQ format. The resulting FASTQ files were then subjected to quality control by trimming the first and last 30 base calls (low quality) and additionally filtering for base calls with PHRED quality score of 15 (>97% accuracy) or better. Sequences with<40% of the remaining bases passing the quality filter were filtered out from the analysis. After quality control of the sequences, multiple sequence alignment (MSA) was carried out. In the MSA, the sample sequences (total of 5), the unique set of top three blast hits (17 sequences), and the downloaded sequences from the genus Photorhabdus (3 sequences) were aligned using the L-INS-i algorithm implemented in the MAFFT tool. The resulting alignment was further trimmed to remove gapped regions using the tool TRIMAL, and the resulting alignments in FASTA format were converted to PHYLIP format using a custom script based on Biopython.
Molecular Identification using 16S rRNA gene
The sequences of a 16S rRNA gene region amplified from three bacterial samples were obtained. The amplicons were sent to the sequencing laboratory of the Philippine Genome Center (PGC) and had sequenced using 27F and 1492R (Forward and Reverse primer sequences). All sequences that passed the quality control procedure were then converted to FASTA format. All of the sequences that passed the quality control step were used as input to the Basic Local Alignment Search Tool (BLAST). The sequences were queried against the nucleotide database of NCBI GenBank using the BLASTN algorithm. For each query, the top three database hits were recovered and included in the subsequent analysis and three 16S rRNA gene sequences from the genus Photorabdus were also downloaded for the outgroup analysis of the phylogenetic tree.
Phylogenetic tree reconstruction of isolated bioluminescent bacteria
For the phylogenetic tree reconstruction, multiple sequence alignment was carried out as input to reconstruct a phylogenetic tree using the PhyML as a tool, under the K80+I nucleotide substitution model, with 1000 bootstrap replicates. The substitution model was chosen because it has the lowest Bayesian Information Criterion (BIC) for finding the best fit substitution model for the analysis. The resulting maximum likelihood tree was generated initially in Newick format (phyml_tree.txt) and viewed using the FigTree as a tool. The tree was viewed in rectangular orientation.
Morphologically-identified marine crustaceans
The four out of seven marine crustaceans with bioluminescent isolates collected from northeastern section of Manila Bay belonging to order Decapod were identified and verified based on journal website of World Register of Introduced Marine Species (WRiMS) and Global Biodiversity Information Facility (GBIF)26 were Oratosquilla oratoria (O. oratoria)- Mantis Shrimp, Thenus orientalis (T. orientalis) – Oriental Slipper Lobster, Scylla serrata (S. serrata) -Mud Crab and Penaeus monodon (P. monodon)- Giant Tiger Prawn. Figure 1 presents the dorsal and ventral structures of the crustacean specimen, respectively. (A-B) O. oratoria– Mantis Shrimp, (C-D)(T. orientalis) – Oriental Slipper Lobster, (E-F)(S. serrata) -Mud Crab and (G)(P. monodon)- Giant Tiger Prawn.
Figure 1. Four crustaceans species collected from Manila Bay: O. oratoria or Mantis Shrimp Lobster (A1. Dorsal and (A2. Ventral); T. orientalis or Slipper Lobster (B1. Dorsal and B2. Ventral); S. serrata or Mud Crab (C1. Dorsal and; C2. Ventral); P. monodon or Giant Tiger Prawn (D)
Presence of bioluminescent bacterial isolates
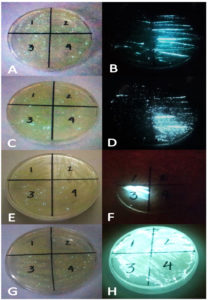
The seven marine crustaceans were swabbed from the inner carapace, eyes, abdomen, abdominal segments and were streaked separately in bioluminescent agar. However, four out of seven marine crustaceans O. oratoria, T. orientalis, S. serrata, and P. monodon contained bioluminescent bacteria which were successfully isolated. Table 1 shows the presence of BLB in the inner surface of carapace and the eyes, the abdomen of O. oratoria, T. orientalis and S. serrata and the abdominal segments of O. oratoria and T. orientalis. On the other hand, Figure 2 shows the first and second isolation of bioluminescent bacteria from four (4) marine crustacean species collected.
Table (1):
Four marine crustaceans collected from Manila Bay that exhibited luminescence and its bacterial colonies; O. oratoria (Mantis shrimp), T. orientalis (Slipper lobster), P. monodon (Giant tiger prawn), S. serrata (Mud crab)
Species |
O. oratoria |
T. orientalis |
P. monodon |
S. serrata |
|---|---|---|---|---|
Inner surface of carapace |
+ |
+ |
+ |
+ |
Eyes |
+ |
+ |
+ |
+ |
Abdomen |
+ |
+ |
– |
+ |
Abdominal segments |
+ |
+ |
– |
– |
*(+) Present; (-) Not present luminescent bacterial colonies
Figure 2. The first and second isolation of bioluminescent bacteria from four of the crustacean species collected from Manila Bay: O. oratoria or Mantis Shrimp Lobster (A. First isolation and B. Second); T. orientalis or Slipper Lobster (C. First isolation and D. Second); S. serrata or Mud crab (E. First isolation and F. Second); P. monodon or Giant Tiger Prawn (G. First isolation and H. Second)
Molecular-based identification of bioluminescent bacterial isolates
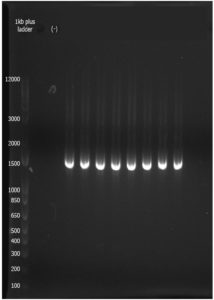
The isolated bacteria from the parts of the four crustacean species were upscaled; however, the BLB from S. serrata showed no visible pellet, hence was not extracted. The genomic DNA (gDNA) isolation followed the protocols from QIAPrep® Spin Miniprep Kit. A 3 µl of the resolved genomic DNA was analyzed by electrophoresis on a 1.2% agarose gel in 100V for 40mins with DNA marker 1kb plus ladder (Invitrogen). All samples showed partially degraded gDNA with thick smeared bands from 1,000bp to ~12,000bp. Isolated gDNA were subjected to gene amplification by polymerase chain reaction (PCR). Components include genomic DNA, 27F and 1492R universal 16S primers, Taq buffer, DNA polymerase, and dNTP mix. Amplicons exhibited intense band smear for each sample—loaded to 1.2% agarose gel run in 100V for 35 minutes DNA marker 1kb plus ladder (Invitrogen). The AGE of amplified 16s rRNA gene showed with region ~1,500bp size (Figure 3). Bioluminescent bacterial strains with complete isolation were processed for the identification based on amplified 16S rRNA gene sequence with an average length of ~770bp size. Based on the analysis, BLAST hits of the five 16S rRNA gene sequences found from the genera of Vibrio alginolyticus and Vibrio rumoiensis.
Bioluminescence is a phenomenon that is widely observed among the diverse groups of organisms from bacteria to vertebrates.10,12 One of the largest groups of bioluminescent bacteria is the genera of Vibrio which is composed of greater than 100 species clustered into 14 clades under the family of Vibrionaceae that are naturally residing in fresh and saline aquatic environments. Approximately 12 out of >100 known Vibrio spp. cause infections to humans, infections are normally acquired through exposure to seawater or consumption of undercooked or raw seafoods.28 For several years, vibrio disease outbreaks create an impact on the economic, personal, and public health. The risk factor of consuming raw or undercooked seafood and shellfish exposed to natural environments are strongly associated with diarrhea, and other intestinal infections.29, 30 In the Philippines, the most common Vibrio species are V. cholerae, known for its disease-causing outbreak due to consumption of water or undercooked or raw meat and seafood products, followed by V. parahaemolyticus, V. alginolyticus, V. harveyii, and V. anguillarium which are commonly found in marine organisms.4,6,31
Vibriosis is one of the major disease problems in aquaculture that resulted in high mortality and severe economic loss in the production of shrimps and other crustacean species. The most common causing species are V. alginolyticus, V. harveyii, V. anguillarium, and V. parahaemolyticus.4,31 From the two-year (1983-1984) survey of etiologic survey of diarrheal disease conducted at Manila, Philippines, the prevalence of Vibrio pathogens in 2,908 patients with diarrhea admitted was determined: 3.8% V. cholerae biotype eltor; 2.8% non-OI V. cholerae; 1.7% V. parahaemolyticus; 1.1% other Vibrio spp.6Aquaculture and fisheries in the Philippines had secured the contribution of food, employment, and foreign exchange. P. monodon, together with the S. serrata are the major aquaculture species in the Philippines. This is growing much faster than the capture fisheries. In the early 1990s, due to the over production of P. monodon using high stocking densities these had led to disease outbreak and mass mortalities, which is mainly caused by luminous vibriosis.32
In this study, the bioluminescent bacteria V. alginolyticus, Mucus bacterium, and V. rumoiensis were isolated from the four crustacean species, O. oratoria, T. orientalis, S. serrata, and P. monodon, respectively. They are mostly observed in the eyes and inner carapace (head region) of all host organisms. Vibrionaceae symbioses are widespread in most marine organisms, whether it is mutualism, host-pathogen relationship, or commensalism. Some Vibrio spp. is known for its blue or green-light bioluminescence. The presence of these microorganisms in crustaceans’ eyes indicates a symbiotic interaction between the symbiont and its host, which is employed for communication, prey capture, and attracting mates.9,10,14,16 In most cases, these pathogenic strains of Vibrio species had caused mass mortalities on the marine organisms that led to countless economic losses and risk to the public health.4,5 These bioluminescent bacteria are not only harmful to both human and animal, in other cases, these could be useful for various applications, medically and environmentally. For biomedical application, bioluminescence in bacteria is achieved through the regulation of the oxidative enzymes called luciferase. These enzymes could induce to other bacteria to produce light signal for the bioluminescence imaging used to detect infections, malignancies and cancers.20-23 Secondary metabolites of non-pathogenic Vibrio sp. also have the potential of combating deadly pathogens including Staphylococcus aureus and Klebsiella pneumoniae, which could greatly contribute to the medical field in the Philippines.24 Due to the high sensitivity of bioluminescent bacteria, it could be used in detection of heavy metals and toxic substances present in food and water. For instance, heavy metals from the abandoned mining area have been distributed to the landfill and water sources of the residence in the Philippine Island of Western Mindanao. Early detection of heavy metal present in water and food sources including mercury could possibly prevent the mass poisoning or complications in the community.18,19,25
Bioluminescent microorganisms could also serve as bioindicator species on the totality of marine ecosystems.33 Pathogenic Vibrio spp. were isolated from the Pernaviridis (Green Mussels) in Bacoor bay, a large inlet of south-eastern Manila Bay, the isolates include V. alginolyticus, V. cholerae, V. parahaemolyticus, and V. vulnificus. Manila bay marine ecosystem is highly contaminated due to the presence of pathogenic Vibrio species that may affect the development of juvenile crustacean species and other marine organisms which could severely affect the production of seafood leading to economic loss and danger to public health.34
Approximately 1,500bp size of the 16S rRNA gene was amplified from all the marine crustaceans’ bioluminescent isolates; indicates a positive relevance of genomic DNA to bioluminescent bacteria with 1,500bp size. In this study, it was determined that the bacteria isolated from the four crustacean species belongs to the genera of Vibrio. The 16s rRNA gene sequences of the V. alginolyticus isolated from the eyes and inner carapace region of O. oratoria and T. orientalis showed significant hits: 95.010%, 98.159%, and 97.917%, respectively (Table 2). Molecular identification of Vibrio species, including V. alginolyticus has been studied and still need exploration for possible intervention in its pathogenesis especially in aquaculture settings. In the study of Xue et al.,35 using a thorough pangenome analysis, the genomes of seven V. alginolyticus strains isolated from shrimp larviculture ponds in four provinces of Southeast China were thoroughly characterized. The toxicity of the seven isolates to shrimp postlarvae and their chitin utilization were also determined. These findings may aid in the development and prevention of vibriosis in penaeid shrimp by a thorough examination of the phylogeny and genetic composition of various V. alginolyticus strains, along with their pathogenicity and chitinolytic capacity.
Table (2):
Significant hits of each bioluminescent bacterial isolates from each species of crustacean and their homologies based on BLASTN with the given accession number
Crustacean Species |
Parts of isolation |
Identity (%) |
Orthologues |
Accession Number |
|---|---|---|---|---|
O. oratoria |
inner carapace |
95.010%, |
V. alginolyticus |
MH611367.1 |
T. orientalis |
inner carapace |
98.159% |
V. alginolyticus |
MK167375.1 |
T. orientalis |
eyes |
97.917% |
V. alginolyticus |
MH643643.1 |
P. monodon |
inner carapace |
98.082% |
Mucus bacterium |
AY654787.1 |
P. monodon |
eyes |
97.945% |
V. rumoiensis |
MH712966.1 |
V. alginolyticus is a halophilic pathogenic bacterium which is formerly known as biotype 2 of V. parahaemolyticus that could be acquired by exposure to contaminated salt water and consumption of raw or undercooked seafood. Pathogenic V. alginolyticus can cause severe infections to different crustacean species, as well as in humans. In marine species, pathogenic Vibrio rate of virulence and mortality are mainly higher to the crustacean’s larval stage due to their ability to produce exotoxins which invade the larval tissues causing necrosis that could lead to mass mortalities. This pathogenic vibrio species also devastated the P. monodon and shell clam (Ruditapes decussatus) hatcheries in the Philippine water that resulted in a mass mortality of juveniles.7,36 Aside from P. monodon as crustacean host for V. alginolyticus, a study was carried out in Penaeus vannamei (White leg shrimp). High-throughput sequencing was used to study the apoptosis and autophagy involved in the white leg shrimp’s response to bacterial infection by identifying miRNA-mRNA interactions in the haemocyte of Penaeus vannamei after Vibrio alginolyticus injection. As a result of the injection of Vibrio alginolyticus, the DNA damage in Penaeus vannamei hemocytes increased, and immunological reactions such autophagy and apoptosis significantly increased. The identification of mRNAs and miRNAs from Penaeus vannamei infected with V. alginolyticus also utilized high-throughput sequencing methods. After the V. alginolyticus challenge, a substantial number of miRNAs displayed changed expression, and the majority of them were connected to immunity.37
Extremely potent poison known as TTX or Tetrodotoxin are originally found in the ovary and liver of puffer fish are mainly the toxin produced by the marine bacteria V. alginolyticus and other Vibrio spp.38 TTX is a potent neurotoxin which blocks voltage-gated sodium channels that causes heart failure, paralysis and consequently death. Due to its paralysis effect, this neurotoxin could be potentially used as an analgesic to treat severe various types of pain.39 The V. alginolyticus isolated from crustaceans in this study can be explored and further studies should be conducted on the isolation of this neurotoxin. Another study conducted regarding V. alginolyticus, they looked at the histological changes in Macrobrachium rosenbergii groups that had received experimental intramuscular injections of 50L of 107 CFU of V. alginolyticus between the second and third abdominal segment. The study also shown the importance of using immunohistochemistry and histology to diagnose vibriosis in giant freshwater prawns in order to develop a successful control and treatment plan.40
In Bangladesh, a study conducted to detect the presence of antimicrobial-resistant Vibrio species. The findings supported thorough studies on the prevalence and antimicrobial resistance profiles of Vibrio species, particularly isolates of V. cholerae, V. parahaemolyticus, and V. alginolyticus derived from shrimp and shrimp habitats. This shows that antimicrobial-resistant Vibrio species are present in shrimp and shrimp settings, necessitating a thorough surveillance program including numerous farms and samples from all across the country.41 In non-crustacean marine organisms, an outbreak of vibriosis characterized by serious ulcers in the skin was reported in cultured seahorses. It emphasizes the role of Vibrio harveyi and Vibrio alginolyticus in the disease outbreak in the aquaculture of farmed seahorses and emphasizes the significance of creating early diagnostics and suitable prevention measures to lessen the harm caused by Vibrio spp. in a setup for seahorse farming.42
Another disease caused by Vibrio spp. are extraintestinal, ear, and wound infections when exposed to V. alginolyticus, clinical features may include diarrhea, otitis, mild cellulitis and seropurulent exudate.43 Bacterial isolates from the inner carapace region of P. monodon showed a significant hit of 98.082% with 16s ribosomal rRNA gene sequence of Mucus bacterium. Isolates from the eyes of the P. monodon showed a significant hit of 97.945% (Table 2) with 16S ribosomal rRNA gene sequence of V. rumoiensis, a rod-shaped and non-flagellated microorganism with numerous blebs on its surface. Presence of these blebs on its surface could be associated with its ability to grow in a H2O2-containing environment. The V. rumoiensis isolated from the eyes of P. monodon have a symbiotic relationship to thrive in a H2O2-containing environment. The pathogenicity and function of the V. rumoiensisare still unknown; however, this bacterial strain shown a higher catalase activity than other bacterial species such as Escherichia coli, V. parahaemolyticus, and Micrococcus luteus, which are well known for their high catalase activity.44
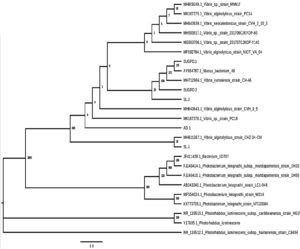
Figure 4. Maximum likelihood phylogenetic tree of the 16S rRNA gene of the isolated bioluminescent bacteria. The tree was inferred from 16S rRNA gene sequences using MEGA 10 with a 1000 bootstrap replicates. Sample ID were named on the following in the phylogenetic tree (SUGPO.1 – Mucus bacterium from P. monodon host, SUGPO.2 – Vibrio rumoiensis strain from P. monodon host, SL.2 – Vibrio alginolyticus strain from T. orientalis host, AD.1 – Vibrio alginolyticus strain from O. oratoria host, SL.1 – Vibrio alginolyticus strain from T. orientalis host)
The phylogenetic tree reconstruction analysis of the bacterial isolates illustrates a clear separation of the genera under the Family Vibrionaceae in correlation of their hosts and habitats where they are usually observed (Figure 4). The class of Proteobacteria including the genera of Vibrio, Photobacterium, and Photorhabdus were genera of bacteria known to produce bioluminescence or green light.8-10,16 The constructed phylogenetic tree shows seven major clades. Clade 1 includes Vibrio sp., V. alginolyticus, and V. neocaledonicus strains. These bacterial strains are commonly found in different marine organisms such as fish, corals, sea urchins, and crustaceans.2 The phylogenetic analysis of the three bacterial isolates Mucus bacterium, V. rumoiensis,and V. alginolyticus which are respectively isolated from P. monodon and T. orientalis host, are existed in the Clade 2 which showed a closer phylogenetic relatedness. This could highly be expected since V. alginolyticus has been considered as a pathogenic bacterium of some marine crustacean species, including P. monodon.7 Based on the 16S rRNA gene sequences of the bacterial isolate, Mucus bacterium, acquired from the NCBI database of GenBank, this isolate was taken from the mucus DNA of Oculinapatagonica. Based on the mucus DNA analysis, the most abundant bacteria present in the mucus of O. patagonica was V. splendidus from winter and summer.45 The bacterial isolates V. alginolyticus from the host of O. oratoria and T. orientalis has two distinct clades; Clade 3 and 4, respectively. The three 16S rRNA gene sequences from the genus Photorabdus used as an outgroup for the analysis of phylogenetic tree.
The present study through molecular identification and phylogenetic tree reconstruction revealed that the isolated bioluminescent symbionts in the Philippine marine crustacean hosts were closely related to the Family Vibrionaceae—V. alginolyticus, V. rumoiensis and Mucus bacterium. This highlights the evidence of the presence of bioluminescent symbionts in the Philippine marine crustacean hosts. This report presents different bioluminescent bacterial species present in crustaceans and would be of great help in the formulation of techniques for conserving the marine species inhabiting the Manila Bay. The Philippine marine crustaceans were established as bioluminescent symbionts. This study will contribute to the very limited knowledge of the great diversity of microorganisms in the enormous marine organisms thriving in the Philippine marine ecosystem and provide greater knowledge for future researchers about various sources of bioluminescent species that are significant for medical and environmental applications.
ACKNOWLEDGMENTS
The authors would like to express their sincere gratitude to the people that led to the completion of the study and their Alma Mater, Bulacan State University and to Angeles University Foundation Integrated School, where most of the laboratory work was conducted.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
NCS, MEBC, and JDF conceptualized the study and conducted the experiment. NCS, MEBC, and JDF wrote original draft. NCAS, MEBC, JDF and CJNO revised original draft. NCS, MEBC, JDF and CJNO wrote, reviewed and edited the manuscript. RFJ, ORA, AJM and CJNO performed supervision. All authors read and approved the final manuscript for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
- Mizan, MFR, Jahid IK, Ha SD. Microbial bio lms in seafood:a food-hygiene challenge. Food Microbiol. 2015;49:41-55.
Crossref - Soto W, Lostroh CP, Nishiguchi, MK. Physiological responses to stress in the Vibrionaceae. Symbioses and stress. Cellular Origin, Life in Extreme Habitats and Astrobiology. (J. Seckback, M. Grube, eds.), Springer, Dordrecht. 2010
Crossref - Raissy M, Moumeni M, Ansari M, Rahimi E. Occurrence of Vibrio spp. in lobster and crab from the Persian Gulf. J Food Saf. 2012;32(2):198-203.
Crossref - de la Pena, LD, Lavilla-Pitogo CR, Paner MG. Luminescent Vibrios associated with mortality in pond-cultured shrimp Penaeusmonodon in the Philippines. Fish Pathology. 2001;36(3):133-138.
Crossref - Caipang CMA, Avenido P, Dechavez R, Jaspe CJ. Moderate inhibition of luminous Vibrio harveyii by aqueous extracts obtained from the skin of Tilapia, Oreochromis sp. Philipp J Sci. 2011;140 (2):173-178.
- Adkins HJ, Escamilla J, Santiago LT, Ranoa C, Echeverria P, Cross JH. Two-year survey of etiologic agents of diarrheal disease at San Lazaro Hospital, Manila, Republic of the Philippines. J Clin Microbiol. 1987;25(7):1143-1147.
Crossref - Lavilla-Pitogo CR, Baticados MCL, Cruz-Lacierda ER, de la Pena LD. Occurrence of luminous bacterial disease of Penaeusmonodon larvae in the Philippines. Aquaculture. 1990;91(1-2);1-13.
Crossref - Haddock SH, Moline MA, Case JF. Bioluminescence in the sea. Ann Rev Mar Sci. 2010;2:443-493.
Crossref - Rios-Velazquez C, Malave-Orengo J, Rubio-Marrero E. Isolation and characterization of bioluminescent bacteria from marine environments of Puerto Rico. Current Research, Technology and Education Topic Applied Microbiology and Microbial Technology. 2010;1:103-108
- Widder EA. Bioluminescence in the ocean:origins of biological, chemical, and ecological diversity. Science. 2010;328:704-708.
Crossref - Jia K, Ionescu RE. Measurement of bacterial bioluminescence intensity and spectrum:Current physical techniques and principles. Adv Biochem Eng Biotechnol. 2016;154:19-45.
Crossref - Zarubin M, Belkin S, Ionescu M, Genin A. Bacterial bioluminescence as a lure for marine zooplankton and fish. Proc Natl Acad Sci USA. 2012;109(3):853-857.
Crossref - Naguit MAA, Plata KC, Abisado RG, Calugay RJ. Evidence of bacterial bioluminescence in a Philippine squid and octopus hosts. ACCL Bioflux. 2014;7:497-507.
- Iqbal BMM, Rajendran S, Vasudevan S. Isolation, identification and characterization of the bioluminescent bacteria isolated from the blue swimmer crab Portunus pelagicus along Thondi Coast and virulence studies at high temperatures. Microb Pathog. 2018;117:232-236
Crossref - Cronin M, Akin AR, Collins SA, et al. High Resolution in vivo bioluminescent imaging for the study of bacterial tumour targeting. PLoS One. 2012;7:e30940.
Crossref - Wannamaker AM. Quorum sensing in vibrios and cross-species activation of bioluminescence lux genes by Vibrio harveyiLuxR in an arabinose-inducible Escherichia coli expression system. M.S thesis, University of Wisconsin Milwaukee, Wisconsin, United States. 2013
- Arante LAB, Sison JAD, Molina JA. A report on bioluminescent bacteria symbiont from Philippine marine fishes. Research Journal of Fisheries and Hydrobiology. 2015;10(16):1-4.
- Hong Y, Chen Z, Zhang B, Zhai Q. Isolation of Photobacterium sp. LuB-1 and its application in rapid assays for chemical toxicants in water. Lett Appl Microbiol. 2010;51(3):308-312.
Crossref - Molina A, Tuazon R, Larman L. Responses of bioluminescent bacteria isolated from Philippine marine fishes to various heavy metals. Aust J Basic Appl Sci. 2015;9:38-42.
- Kong Y, Shi Y, Chang M, et al. Whole-body imaging of infection using bioluminescence. Curr Protoc Microbiol. 2011.
Crossref - Chang MH, Cirillo SL, Cirillo JD. Using luciferase to image bacterial infections in mice. J Vis Exp. 2011;48:2547.
Crossref - Gonzalez RJ, Weening EH, Frothingham R, Sempowski GD, Miller VL. Bioluminescence imaging to track bacterial dissemination of Yersinia pestis using different routes of infection in mice. BMC Microbiol. 2012;12:147.
Crossref - Dworsky EM, Hedge V, Loftin AH, et al. Novel in vivo mouse model of implant related spine infection. J Orthop Res. 2017;35(1):193-199.
Crossref - Molina AJ, Abisado RG, Calugay RJ. Bioluminescent Vibrio spp. with antibacterial activity against the nosocomial pathogens Staphylococcus aureus and Klebsiella pneumoniae. ACCL Bioflux. 2016;9(2):185-194.
- Cortes-Maramba N, Reyes JP, Francisco-Rivera AT, Akagi H, Sunio R, Panganiban LC. Health and environmental assessment of mercury exposure in a gold mining community in Western Mindanao, Philippines. J Environ Manag. 2006;81(2):126-134
Crossref - Rius M, Ahyong S, Costello MJ, et al. World Register of Introduced Marine Species. (WRiMS). 2021.
Crossref - Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173 (2):697-703.
Crossref - Baker-Austin C, Oliver JD, Alam M, et al. Vibrio spp. infections. Nat Rev Dis Primers. 2018;4(8); 1-19.
Crossref - West PA. The human pathogenic vibrios- a public health update with environmental perspectives. Epidemiol Infect. 1989;103(1):1-34.
Crossref - Osunla CA, Okoh AI. Vibrio pathogens:A public health concern in rural water resources in sub-saharan Africa. Int J Environ Res Public Health. 2017;14(10):1188.
Crossref - Kumaran T, Citarasu T. Isolation and characterization of Vibrio species from shrimp and Artemia culture and evaluation of the potential virulence factor. Intellectual Property Rights. 2016;4(1):153.
Crossref - Food and Agriculture Organization (FAO). Fisheries and Aquaculture. Philippines. National Aquaculture Sector Overview Fact Sheets. Text by Paclibre, JO In:FAO Fisheries Division. www.fao.org/fishery/countrysector/naso_philippines/en, Accessed 6 Jun 2021
- Kratasyuk VA, Kudryasheva NS, Kendogina EV, Vetrova EV, Kudinova IY. Development of the bioluminescent bioindicators for analyses of pollutions. Field Screening Europe. Springer, Dordrecht 1997:207-210.
Crossref - Tabo NA, Ramirez VB, Tabo HAL, Gloriani NG. Occurrence and antimicrobial resistance of pathogenic vibrios isolated from green mussel, Pernaviridis L. 1758 in Bacoor bay, Cavite, Philippines. Acta Medica Philippina. 2015;49(4):39-49.
Crossref - Xue M, Huang X, Xue J, et al. Comparative Genomic Analysis of Seven Vibrio alginolyticus Strains Isolated from Shrimp Larviculture Water With Emphasis on Chitin Utilization. Front Microbiol. 2022;13, 925747.
Crossref - Gomez-Leon J, Villamil L, Lemos ML, Novoa B, Figueras A. Isolation of Vibrio alginolyticus and Vibrio splendidus from aquacultured carpet shell clam (Ruditapes decussatus) larvae associated with mass mortalities. Appl Environ Microbiol. 2005;71(1):98-104.
Crossref - Wang F, Huang L, Liao M, et al. Integrative analysis of the miRNA-mRNA regulation network in hemocytes of Penaeusvannamei following Vibrio alginolyticus infection. Dev Comp Immunol. 2022;131:104390.
Crossref - Narashi T. Tetrodoxin. Encyclopedia of Neurological Sciences (Second Edition), Academic Press (MJ Aminoff& RB Daroff). 2014;420-422.
Crossref - Lago J, Rodriguez LP, Blanco L, Vieites JM, Cabado AG. Tetrodotxin, an extremely potent marine neurotoxin:distribution, toxicity, origin, and therapeutical uses. Mar Drugs. 2015;13(10):6384-6406.
Crossref - Ajadi A, Sabri MY, Atata JA, Daodu OB, Emikpe BO. Pathology and immunohistochemical evaluation of Vibrio alginolyticus infection in Macrobrachiumrosenbergii. Comp Clin Pathol. 2019;359-368.
Crossref - Haque ZF, Islam MS, Sabuj AAM, et al. Molecular Detection and Antibiotic Resistance of Vibrio cholerae, Vibrio parahaemolyticus, and Vibrio alginolyticus from Shrimp (Penaeus monodon) and Shrimp Environments in Bangladesh. Aquaculture Research. 2023.
Crossref - Xie J, Bu L, Jin S, et al. Outbreak of vibriosis caused by Vibrio harveyi and Vibrio alginolyticus in farmed seahorse Hippocampus kuda in China. Aquaculture. 2020;523:735168.
Crossref - Chart H. Vibrio, mobiluncus, gardnerella and spirillum:Cholera; vaginosis; rat bite fever. Medical Microbiology (Eighteenth Edition) (D. Greenwood, M. Barer, R. Slack, & W. Irving, eds.).2012;314-323. Churchill Livingstone.
Crossref - Yumoto I, Iwata H, Sawabe T, et al. Characterization of facultatively psychrophilic bacterium, Vibrio rumoiensis sp. nov., that exhibit high catalase activity. Appl Environ Microbiol. 1999;65(1):67-72.
Crossref - Koren O, Rosenberg E. Bacteria associated with mucus and tissues of the coral Oculina patagonica in summer and winter. App Environ Microbiol. 2006;72(8):5254-5259.
Crossref
© The Author(s) 2023. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.