ISSN: 0973-7510
E-ISSN: 2581-690X
A total of seventy-two bacterial isolates were obtained employing enrichment culture technique from apple orchard soils contaminated with chlorpyrifos. Pure cultures of bacterial isolates were obtained using streak plate method on mineral salt medium. Bacterial isolates were characterized on the basis of morphology, culture and biochemical properties. Six bacterial isolates exhibited high extracellular organophosphorus hydrolase activity along with high tolerance towards high concentrations of chlorpyrifos. Genomic DNA extraction from bacterial isolates was done with phenol/chloroform method. Molecular Diversity of six chlorpyrifos degrading bacterial isolates was done employing RAPD-PCR technique by using 25 decamer primers, where amplification was showed by only 20 primers. A total of 337 amplified bands and 64 unique bands ranging in size from 100 to 4900bps were produced after RAPD analysis. The similarity coefficient estimated by Jaccard’s coefficient for these bacterial isolates was found to range between 31 to 64 percent.
Chlorpyrifos, Enrichment Technique, Molecular Diversity, Organophosphorus hydrolase (OPH), Random amplification of polymorphic DNA (RAPD)
Organophosphate pesticides (OPs) are used excessively in agriculture and nearly constitute 38 percent of total pesticides usage worldwide1.OPs are reported to be biodegradable in nature but at the same time they are toxic to mammals as well as non-target vertebrates & non-vertebrates. Chlorpyrifos is an example of organophosphate pesticide which is found to be very effective against broad spectrum of insect pests because chlorpyrifos is capable of irreversibly inhibiting acetylcholinesterase (AChE) enzyme in these pests2. Chlorpyrifos is used by farmers as formulations such as emulsifiable concentrates (ECs), granules, wettable powder in apple orchards to control insect pests like wooly apple aphids, thrips, root borer, spider mites etc. Indiscriminate usage of chlorpyrifos in apple orchards by farmers has contaminated soil, air, ground as well as surface water bodies and it has been reported by many researchers that half-life of chlorpyrifos ranges from few days to even years in soil. Farmer’s uses back-pack reservoirs for pesticide application andhave low safety knowledge due to which they are at high risk of exposure to pesticides.
Agrochemicals and their degradation products ultimately remains in soil and it has been reported that only a small percentage of total quantity of agrochemical used is actually involved in the control of pests and diseases.Due to the presence of agrochemical in soils, microbial population adapts and sometimes also develops enzyme systems which help them to rapidly metabolize and consequently eliminate agrochemical from the environment3. Degradation as well as tolerance for contaminant in microbes is observed when they are present in pesticide contaminated sites for long duration4.If soil microbes start using pesticide as a carbon and energy sources, then they can easily degrade or detoxify the pesticide. Microbes with metabolic pathways for degradation of chlorpyrifos have been considered as the cost effective and environmental friendly approach for the bioremediation of chlorpyrifos contaminated ecosystems. Chlorpyrifos has been reported to degraded by various bacteria belonging to genera Pseudomonas4,5,6,7, Enterobacter8, Bacillus9,10, Klebsiella11, Paracoccus12,, Flavobacterium14,Kocuria15,16, Staphylococcus17, Mesorhizobium.18
In the present study our aim was to isolate bacterial isolates from apple orchard soils with potential to breakdown chlorpyrifos and study microbial diversityamong chlorpyrifos degrading bacteria using RAPD-PCR technique.
Sample collection
Collection of the soil samples were done from pesticide contaminated soil of apple orchards from Kotkhai, Matiyana, Theog, Chopal and Rohru sites of district Shimla, Himachal Pradesh. Soil samples in triplicates using sterile spatula were collected in sterilized polythene bag from the depth of 10 – 15 cm and then were stored at 4°C in refrigerator. Chlorpyrifos (O, O-diethyl O-(3, 5, 6-trichloro-2-pyridyl) phosphorothioate) with 99.6% purity was procured from Sigma – Aldrich, Switzerland.
Isolation of chlorpyrifos degrading bacteria by enrichment technique
The soil samples were used for enrichment after air drying at room temperature and sieving by using 2mm mesh. The 10 gm of soil mixed with 50 ml of modified minimal salt medium (MSM) broth19 (gl-1: K2HPO4 0.5g, KH2PO4 0.5 g, (NH4)2SO4 0.5 g, NaCl 0.5 g, MgSO4 0.2 g, CaCl2 0.05g, FeSO4 0.02)19containing 10 mg/l of chlorpyrifos and incubated at 37°C under dark condition on orbital shaker for a week duration. Following incubation period, transferred 10 ml of this one-week old culture in fresh 50 ml of MSM broth containing 30mg/l of chlorpyrifos and again incubated for a week on shaker. Further transfer 10 ml of this culture into the fresh 50 ml MSM containing 50mg/l chlorpyrifos and incubated in same conditions for a week. Chlorpyrifos degrading and resistant bacteria were isolated by spread plate and streak plate technique by using this enriched culture.6,20
Screening of chlorpyrifos degrading bacteria
High chlorpyrifos tolerance and degradation
Isolates were screened on eosin-methylene blue agar (EMBA) indicator medium fortified with50mg/l chlorpyrifosand incubated at 37°C for 48 hrsto observe color change in of the colony into red21. Further, each bacterial isolate with chlorpyrifos degrading activity was streaked on MSM agar with varying concentration of chlorpyrifos (50-800 mg/l).22,23
Organophosphorus hydrolase (OPH) activity
Bacterial isolates with high chlorpyrifos tolerance and degrading abilities were then screened for Organophosphorus hydrolase activity. Overnight culture of bacterial isolates in Luria Bertani (LB) medium was centrifuged at 8,000 x g, 4°C for 10 minutes and further supernatant was treated as crude extracellular enzyme. Resuspend cell pellet in 50 mmol/l Tris-Cl (pH 8.0) buffer containing 0.1 mmol/l phenylmethylsulphonyl fluoride (PMSF) after washing twice. Cell disruption was done using sonication 10 times for 10 seconds with interval of 15 seconds (Digital Sonifier-MP Biomedical). Intact cells were separated by centrifugation at 15000 × g for 30 minutes following collection of supernatant which further was used for quantitative enzyme assay. For quantitative enzyme assay, standard method was used24.
Assessing molecular genetic diversity of chlorpyrifos degrading bacteria
Genomic DNA extraction and quantification
Standard method using phenol-chloroform-isoamyl alcohol (25:24:1) was employed for total genomic DNA extraction from the selected isolates and further DNA precipitation was done with isopropanol. This was followed by treating extracted DNA with RNase (0.2 mg/ml) at 37°C for 15 min, and then again phenol-chloroform-isoamyl alcohol was used for extraction followed by isopropanol precipitation. DNA pellet was then re-suspended in Tris EDTA buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0), stored at -20°C, till further use as template DNA for RAPD-PCR.25Quantification of the extracted DNA samples were done on 1.0% agarose gel, in 1X Tris-acetate EDTA (TAE) buffer (2M Tris base, 57.10 ml acetic acid and 0.5 M EDTA (pH 8.0, 50X) comprising ethidium bromide (0.5 µg/ml). DNA samples were mixed with loading dye and load in a well of gel and allowed to get separated at 80V for 1.5-2 hours using power system. Gel Documentation system was employed for visualization and capturing the gel image.
RAPD – PCR analysis
Genomic DNA from the bacteria were analyzed by modified RAPD method of Williams 26.The RAPD analysis was done with 25 primers (13 OPA, 10 OPB and 02 OPC series) supplied by Xcelris labs, India by using thermocycler. PCR amplification was performed with 20µl reaction mixture comprising 13.5 µl deionized water, 2.0µl of 10 X PCR buffer containing MgSO4, 0.5µl of 1U thermo tolerant enzyme (Taq DNA polymerase), 2.0µl of 10 mM dNTPs, 1.0µl of random primer and 1.0µl of template DNA. PCR tubes containing mixture were loaded in the thermocycler for amplification. The PCR protocol comprised of initial denaturation at 94°C for 5 minutes; followed by denaturation at 94°C for 45 seconds and then annealing at 35°C for 1minute followed by extension at 72°C for 1.5minutes and final extension at temperature of 72°C for 8 minutes with 35 number of cycles with final holding at 4°C. Usingagarose gel electrophoresis, amplified PCR products were further analyzed where 1.0% agarose gelwas dissolved in TAE buffer containing 242g ofTris base, Tris, 57.1ml of glacial acetic acid, 100ml 0.5M EDTA with pH 8.0, ethidium bromide (0.5 µg/ml) along with a size DNA ladder of 5 kb and gels were further visualized by means of gel documentation system.Scoring for presence and absence of bands on gels were done for the amplified products of selected bacterial isolates. A score of ‘1’ was specified for presence and ‘0’ was marked for absence of an amplified product. Genetic similarity from the data generated was analyzed using un-weighted pair group arithmetic mean (UPGMA) program of NTSYS-Pc (version 2.02) (Numerical Taxonomy System, Exeter Software). The dendrogram was constructed27 and it serves a base for genetic relatedness between chlorpyrifos degrading bacterial isolates.
In the present study soil samples were collected in triplicates from top soil of apple orchards of Himachal Pradesh. By employing the enrichment culture technique, 72 potential chlorpyrifos degrading bacteria were isolated using mineral salt medium (MSM) which was a modified media previously used for isolation of bacteria19.In literature many researchers till date have reported chlorpyrifos degrading bacteria which were isolated a variety of different sources such as agriculture soil,7,28rhizospheric soils,29 agricultural wastewater,10 sludge and waste water.30,31
It was observed that out of 72 isolates only 15 were able to grow on selective eosin methylene blue agar (EMBA) fortified with 50mg/l chlorpyrifos and color of colonies changed into red, which was considered as indicator for the presence of chlorpyrifos degrading ability. Morphological characterization was followed by biochemical characterization of these 15 bacterial isolates indicated that all of them belong to either g-Proteobacteria or Bacillus genera using Bergey’s manual of determinative bacteriology. Among these fifteen isolates, only six bacterial isolates designated as S1K3.2, S2R1.1, S3T2.1, S3T2.2, S3T3.2, and S4M1.1 exhibited tolerance for the high concentration of chlorpyrifos (g 400mg/l) which may be due to the presence of high extracellular as well as intracellular organophosphate hydrolase activity in these isolates. Researchers has also reported the high intracellular organophosphate hydrolase activity in the individual isolates as well as in consortium which included Pseudomonas aeruginosa, P.stutzeri, P.putida and Klebsiella sp7. Similarly, scientist has also reported extracellular organophosphate hydrolase activity in Pseudomonas resinovaran sstrain AST2.2 and Pseudomonas indol oxydans strain ASK3.2.32
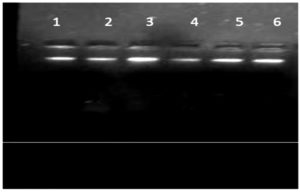
Fig. 1. Agarose gel electrophoresis of genomic DNA extracted from six chlorpyrifos degrading isolates.
Lane 1: S1K3.2 ; Lane 2: S2R1.1 ; Lane 3: S3T2.1 ; Lane 4: S3T2.2 ; Lane 5: S3T3.2 ; Lane 6: S4M1.1
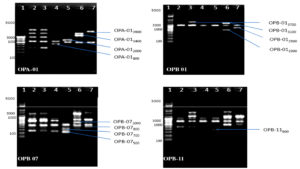
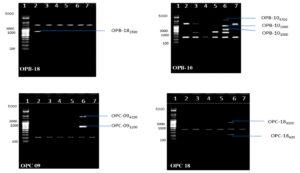
Genomic DNA was extracted from selected six chlorpyrifos degrading bacterial isolates and sharp DNA bands were obtained on 1% agarose gel (Figure 1). Microbial diversity among six bacterial isolates was studied by using RAPD-PCR technique. The PCR amplified products of six selected chlorpyrifos degrading isolates with respect to 25 random primers which were used, where only 20 primers showed 229 polymorphic bands. Five random primers (OPB02, OPB03, OPB05, OPB06 and OPB09) failed in amplification of the genomic DNA in the six isolates. Among 20 random primers which showed amplification 13 belong to OPA series, 5 were from OPB series and 2 were from OPC series. A total of 337 amplified bands ranging in size between 100-4900 bp were recorded after RAPD analysis ofsix chlorpyrifos degrading bacterial isolates.
Table (1):
Twenty five random primer with their nucleotide sequence, total number of bands including unique, monomorphic and polymorphic bands and percent polymorphism.
| No | Primer | Primer Sequence | Isolates |
Total bands | Unique Markers | Monomorphic bands | Polymorphic bands | % Polymorphism | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S1K3.2 | S2R1.1 | S3T2.1 | S3T2.2 | S3T3.2 | S4M1.1 | ||||||||
| 1. | OPA01 | 5’-CAGGCCCTTC-3’ | 5 | 5 | 2 | 2 | 3 | 2 | 19 | 4 | 6 | 13 | 68 |
| 2. | OPA02 | 5’-TGCCGACGTG-3’ | 5 | 4 | 1 | 2 | 4 | 5 | 21 | 6 | 6 | 15 | 71 |
| 3. | OPA04 | 5’-CAGGCCCTTC-3’ | 5 | 4 | 5 | 4 | 6 | 4 | 28 | 5 | 6 | 22 | 79 |
| 4. | OPA06 | 5’-GGTCCCTGAC-3’ | 4 | 4 | 3 | 1 | 4 | 2 | 18 | 2 | 6 | 12 | 67 |
| 5. | OPA07 | 5’-GAAACGGGTG-3’ | 4 | 4 | 3 | 2 | 4 | 5 | 22 | 4 | 0 | 22 | 100 |
| 6. | OPA09 | 5’-GGGTAACGCC-3’ | 2 | 1 | 1 | 1 | 1 | 1 | 7 | 1 | 6 | 1 | 14 |
| 7. | OPA11 | 5’-CAATCGCCGT-3’ | 5 | 5 | 4 | 6 | 5 | 4 | 29 | 2 | 6 | 23 | 79 |
| 8. | OPA12 | 5’-TCGGCGATAG-3’ | 1 | 4 | 2 | 3 | 5 | 8 | 23 | 6 | 6 | 17 | 74 |
| 9. | OPA13 | 5’-CAGCACCCAC-3’ | 5 | 3 | 4 | 6 | 5 | 4 | 27 | 7 | 0 | 27 | 100 |
| 10. | OPA14 | 5’-TCTGTGGCGG-3’ | 1 | 1 | 1 | 1 | 2 | 2 | 8 | 2 | 6 | 2 | 25 |
| 11. | OPA16 | 5’-AGCCAGCGAA-3’ | 1 | 4 | 1 | 1 | 4 | 1 | 12 | 4 | 6 | 6 | 50 |
| 12. | OPA18 | 5’-AGGTGACCGT-3’ | 2 | 1 | 1 | 1 | 1 | 2 | 8 | 2 | 6 | 2 | 25 |
| 13. | OPA20 | 5’-GTTGCGATCC-3’ | 4 | 5 | 1 | 2 | 5 | 5 | 22 | 2 | 6 | 16 | 73 |
| 14. | OPB01 | 5’-GTTTCGCTCC-3’ | 1 | 2 | 1 | 1 | 3 | 2 | 10 | 4 | 6 | 4 | 40 |
| 15. | OPB07 | 5’-GGTGACGCAG-3’ | 5 | 5 | 4 | 3 | 4 | 4 | 25 | 4 | 6 | 19 | 76 |
| 16. | OPB10 | 5’-CTGCTGGGAC-3’ | 2 | 3 | 1 | 2 | 5 | 2 | 15 | 3 | 6 | 9 | 60 |
| 17. | OPB11 | 5’-GTAGACCCGT-3’ | 3 | 4 | 2 | 2 | 5 | 4 | 20 | 1 | 6 | 14 | 70 |
| 18. | OPB18 | 5’-CCACAGCAGT-3’ | 2 | 1 | 1 | 1 | 1 | 1 | 7 | 1 | 6 | 1 | 14 |
| 19. | OPC09 | 5’-CTCACCGTCC-3’ | 1 | 1 | 1 | 1 | 3 | 1 | 8 | 2 | 6 | 2 | 25 |
| 20. | OPC18 | 5’-TGAGTGGGTG-3’ | 1 | 1 | 1 | 1 | 3 | 1 | 8 | 2 | 6 | 2 | 25 |
| 21. | OPB03 | 5’-CATCCCCCTG-3’ | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 22. | OPB06 | 5’-TGCTCTGCCC-3’ | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 23. | OPB02 | 5’-TGATCCCTGG-3’ | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 24. | OPB05 | 5’-TGCGCCCTTC-3’ | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 25. | OPB09 | 5’-TGGGGGACTC-3’ | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Grand total | 337 | 64 | 108 | 229 | 68 | ||||||||
Sixty-four unique bands which can be used as markers for identification of these chlorpyrifos degrading isolates were also obtained (Table 1, Figure 2-6).Researchers has also reported total of 149 polymorphic bands with 20 random primers, while studying molecular diversity of eight chlorpyrifos degrading isolates.29Few researchers has conducted genetic fingerprinting and obtained molecular markers for five bacterial isolates Pseudomonas stuzeri, Enterobacter aerogenes, Pseudomonas pseudoalcaligenes, Pseudomonas maltophila and Pseudomonas vesicularis using RAPD-PCR and PCR-RFLP techniques.33Few DNA fragments were apparently comparable in size among the six bacterial isolates studied, whereas others were unique to individual isolate and further these specific markers can be used for identification as well as tracking them.
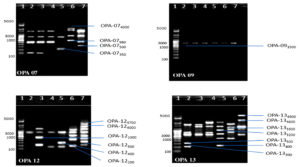
Fig. 2. Banding pattern of RAPD amplified product of six chlorpyrifos degrading isolates by using OPA 7, OPA 9, OPA 12 and OPA 13 primer
Lane 1: 5 kb ladder ; Lane 2: S1K3.2 ; Lane 3: S2R1.1 ; Lane 4: S3T2.1 ; Lane 5: S3T2.2 ; Lane 6: S3T3.2 ; Lane 7: S4M1.1
Fig. 3. Banding pattern of RAPD amplified product of six chlorpyrifos degrading isolates by using OPA 16, OPA 18, OPA 02 and OPA 04 primer
Lane 1: 5 kb ladder ; Lane 2: S1K3.2 ; Lane 3: S2R1.1 ; Lane 4: S3T2.1 ; Lane 5: S3T2.2 ; Lane 6: S3T3.2 ; Lane 7: S4M1.1
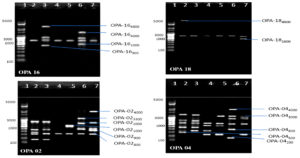
Fig. 4. Banding pattern of RAPD amplified product of six chlorpyrifos degrading isolates by using OPA 06, OPA 11, OPA 14 and OPA 20 primer
Lane 1: 5 kb ladder ; Lane 2: S1K3.2 ; Lane 3: S2R1.1 ; Lane 4: S3T2.1 ; Lane 5: S3T2.2 ; Lane 6: S3T3.2 ; Lane 7: S4M1.1
Fig. 5. Banding pattern of RAPD amplified product of six chlorpyrifos degrading isolates by using OPA 01, OPB 01, OPB 07 and OPB 11 primer
Lane 1: 5 kb ladder ; Lane 2: S1K3.2 ; Lane 3: S2R1.1 ; Lane 4: S3T2.1 ; Lane 5: S3T2.2 ; Lane 6: S3T3.2 ; Lane 7: S4M1.1
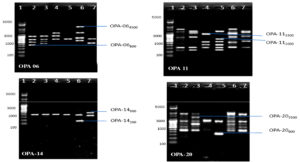
Fig. 6. Banding pattern of RAPD amplified product of six chlorpyrifos degrading isolates by using OPB 18, OPB 10, OPC 09 and OPC 18 primer
Lane 1: 5 kb ladder; Lane 2: S1K3.2 ; Lane 3: S2R1.1; Lane 4: S3T2.1; Lane 5: S3T2.2 ; Lane 6: S3T3.2 ; Lane 7: S4M1.1
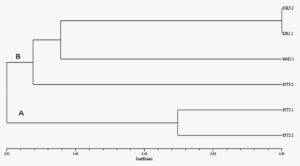
Dendrogram was constructed using UPGMA program where Jaccard’s coefficient was used to estimate the similarities and RAPD-PCR results were also evaluated with SIMQUAL program of NTSYS-Pc (version 2.20). The dendrogram bifurcates into two major clusters which on coefficient scale lies in between 0.31-0.64 (Figure 7). Similar findings were reported with two main clustersin dendrogram lying in coefficient scale range from 0.26 -0.65 and 0.38-1.00 respectively.29, 33 In dendrogram, cluster A comprises of two isolates S3T2.1 and S3T2.2 with 52% similarity with each other whereas cluster B again divided in two sub clusters B1 and B2. In cluster B1 – S3T3.2 isolate shows 34% similarity when compared with other samples in cluster B2. Subsequently, cluster B2 further separated into sub-sub cluster where S1K3.2 and S2R1.1 revealed maximum genetic relatedness of 64%. In the same cluster S4M1.1 showed 38% similarity with the other two isolates S1K3.2 and S2R1.1.
Six bacteria were isolated from apple orchard soils with ability of tolerating high concentrations of chlorpyrifos and they exhibited high extracellular as well as intracellular organophosphorus activities which make them suitable candidate for bioremediation of soils contaminated with chlorpyrifos. RAPD PCR analysis results will be useful for detection of polymorphism and can also establish genetic relatedness among the six bacterial isolates. Identifying as well as tracking of bacterial isolates can be done with unique band which serves as specific marker.
ACKNOWLEDGMENTS
The authors are grateful to Dr. Poonam Shirkot, Professor Biotechnology, Dr. YS Parmar University of Horticulture & Forestry for her help and support during research experiments.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
AKS has conducted the experiments during his Ph.D while KH has edited manuscript and JP designed the figures and tables.
FUNDING
None
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
AVAILABILITY OF DATA
All dataset generated or analyzed during this study are included in the manuscript submitted.
- Singh BK, Walker A. Microbial degradation of organophosphorus compounds. FEMS Microbiol Rev. 2006;30:428-471.
Crossref - Rayu S, Nielsen UN, Nazaries L, Singh BK. Isolation and Molecular Characterization of Novel Chlorpyrifos and 3,5,6- trichloro-2-pyridinol-degrading Bacteria from Sugarcane Farm Soils. Front Microbiol.2017;8:518.
Crossref - Kearney PC, Kaufman DD. Enzymes from soil bacterium hydrolyze phenycarbamate herbicides. Science. 1965;147: 740-741.
Crossref - Farhan M, Khan AU, Wahid A, Ahmad M, Ahmad F. Biodegradation of chlorpyrifos using indigenous Pseudomonas sp. isolated from industrial drain. Pakistan Journal of Nutrition. 2012; 11(12): 1183-1189.
Crossref - Chawla N, Suneja S, Kukreja K. Isolation and characterization of chlorpyriphos degrading bacteria. Indian Journal of Agricultural Research. 2013; 47: 381-391.
- Sharma A, Pandit J, Sharma R, Shirkot P. Biodegradation of chlorpyrifos by pseudomonas resinovarans Strain AST2.2 isolated from enriched cultures. Curr World Environ. 2016;11(1): 267-278.
Crossref - Sasikala C, Jiwal S, Rout P, Ramya M. Biodegradation of chlorpyrifos by bacterial consortium isolated from agriculture soil. World Journal of Microbiology and Biotechnology. 2012; 28: 1301-1308.
Crossref - Singh BK, Walker A, Morgan JAW, Wright DJ. Biodegradation of chlorpyrifos by Enterobacter strain B-14 and its use in bioremediation of contaminated soils. Applied and Environmental Microbiology. 2004; 70(8): 4855-4863.
Crossref - Liu Z, Chen X, Shi Y, Cheng SZ. Bacterial degradation of chlorpyrifos by Bacillus cerus. Advanced Materials Research. 2011;356-360: 676-680.
Crossref - El-Helow ER,Badawy MEI, Mabrouk MEM, Mohamed EAH, El-Beshlawy YM. Biodegradation of Chlorpyrifos by a Newly Isolated Bacillus subtilis Strain, Y242. Bioremediation Journal. 2013; 17(2):113-123.
Crossref - Ghanem I, Orfi M, Shamma M. Biodegradation of chlorpyrifos by Klebsiella sp. isolated from an activated sludge sample of waste water treatment plant in damascus. Folia Microbiol. 2007; 52:423-427.
Crossref - Fan S, Li K, Yan Y, et al. A novel chlorpyrifos hydrolase CPD from Paracoccus sp. TRP: Molecular cloning, characterization and catalytic mechanism. Electronic Journal of Biotechnology. 2018 31:10-16.
Crossref - Xu G, Zheng W, Li Y, Wang S, Jhang J, Yan J. Biodegradation of chlorpyrifos and 3,5,6- trichloro-2-pyridinol by a newly isolated Paracoccussp. strain TRP. International Biodeterioration Biodegradation. 2008; 62: 51-56.
Crossref - Mallick K, Bharati K, Banerji A, Shaki NA, Sethunathan N. Bacterial degradation of chlorpyrifos in pure cultures and in soil. Bull Environ ContamToxicol. 1999; 62:48-54
Crossref - Nagavardhanam N, Vishnuvardhan Z. Isolation, screening and identification of organophosphate pesticide degrading Bacterium, Kocuria sp. Asian Journal of Biological and Life Sciences. 2012; 1: 204-207.
- Hamsavathani V, Aysha OS, Ajith AR. Isolation and identification of chlorpyrifos degrading bacteria from agricultural soil. International Journal of Advance Research. 2017;5(5):1209-1221.
Crossref - Supreeth M, Raju NS. Bio-Mineralization of Organophosphorous Insecticide-Chlorpyrifos and its Hydrolyzed Product 3, 5, 6-Trichloro-2-Pyridinol by Staphylococcus Sp. ES-2. Current World Environment. 2016;11(2).
Crossref - Jabeen H, Iqbal S, Anwar S. Biodegradation of chlorpyrifos and 3, 5, 6 trichloro 2 pyridinol by a novel rhizobial strain Mesorhizobium sp. HN3. Water and Environment Journal. 2015;29(1): 151-60.
Crossref - Yang C, Liu N, Guo X, Qiao C. Cloning of mpd gene from a chlorpyrifos degrading bacterium and use of this strain in bioremediation of contaminated soil. FEMS Microbiology Letters. 2006; 265: 118-125.
Crossref - Harish R, Supreeth M, Chauhan J B. Biodegradation of Organophosphate Pesticide by Soil Fungi. Advanced Biotech. 2013;12:04-08.
- Bhat MA, Tsuda M, Horiike K, Nozaki M, Vaidyananthan CS, Nakazawa T. Identification and characterization of a new plasmid carrying genes for degradation of 2,4- Dichlorophenoxyacetate from Pseudomonas cepacia CSV90. Applied and Environmental Microbiology. 1994; 60: 307-312.
Crossref - Bhagobaty RK, Malik A. Utilization of Chlorpyrifos as a sole source of carbon by bacteria isolated from wastewater irrigated agricultural soils in an industrial area of western Uttar Pradesh, India. Research Journal of Microbiology. 2008; 3(5): 293-307.
Crossref - Sharma AK, Kasture A, Pandit J, Shirkot P. Isolation, characterization and identification of chlorpyrifos tolerant pseudomonas strain exhibiting extracellular organo-phosphorus hydrolase (OPH) activity from apple orchard soils of Himachal Pradesh. Asian Journal of Microbiology, Biotechnology and Environmental Science. 2017; 19(4): 935-944.
- Ningfeng WU, Minjie D, Xiuyun S, Guoyi L, Bin Y, Yunliu F. Isolation, purification and characterization of a new organphosphorus hydrolase OPHC2. Chinese Science Bulletin. 2004; 49(3): 268-272.
Crossref - Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning: A Laboratory Manual. New York, NY: Cold Spring Harbor Laboratory. 1982
- Williams JGK, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Research. 1990; 18: 6531-6535.
Crossref - Rohlf F J. NTSYS- pc: Numerical taxonomy and multivariate analysis system version 1.80. Department of Ecology and Evolution, State University of New York, Stony Brook, NY. 1993.
- Rani MS, Lakshmi KV, Devi PS, et al. Isolation and characterization of a chlorpyrifos degrading bacterium from agricultural soil and its growth response. African Journal of Microbiology Research. 2008;2:26-31.
- Kumar RG, Gopal AV, Reddy RS, Rani CVD,Triveni S, Chari KD. Molecular diversity of organophosphorus degrading bacteria from different field soils. Journal of Pure and Applied Microbiology. 2016; 10(4): 2727-2733.
Crossref - Latifi AM, Khodi S, Mirzaei M, Miresmaeili M, Babavalian H. Isolation and characterization of five chlorpyrifos degrading bacteria. African Journal of Biotechnology. 2011; 11(13): 3140-3146.
Crossref - Li XH, He J, Li SP. Isolation of a chlorpyrifos-degrading bacterium, Sphingomonas sp. strain Dsp-2, and cloning of the mpd gene. Research Microbiology. 2007; 158(2): 143-149.
Crossref - Sharma AK, Pandit J, Gautam V, Shirkot P. Optimization of culture conditions for Maximizing Extracellular Organophosphorus Hydrolase activity in Pseudomonas strains with chlorpyrifos degradation potential. Journal of Pharmacognosy and Phytochemistry. 2019; 8(1): 322-326.
- Awad NS, Sabit HH, Abo-Aba SEM, Bayoumi RA. Isolation, characterization and fingerprinting of some chlorpyrifos- degrading bacterial strains isolated from Egyptian pesticides-polluted soils. African Journal of Microbiology Research. 2011; 5(18): 2855-2862.
Crossref
© The Author(s) 2021. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.