ISSN: 0973-7510
E-ISSN: 2581-690X
Diarrheagenic Escherichia coli still an important pathogen that cause diarrhea which lead to hospital admissions and death specially in children. In order to identify the common pathotypes of E. coli via investigate different virulence genes. A total of 210 stool samples were collected from children under five years presented with diarrhea from different hospitals and private clinics in Wasit province, Iraq, on the other hand, 40 stool samples were collected from healthy children considered as control group. regarding to culture, biochemical tests and API 20E results 100 isolates were supposed to be E. coli. The DNA were extracted to that 100 isolates from diarrheal cases and for 40 isolates of control, concentration of DNA samples were between (50-360 mg/µl ) and the purity between (1.8-2). All isolates studied for detectionvirulence gene of five Diarrheagenic Escherichia Coli strains based on using multiplex Polymerase Chain Reaction technique, by amplified 13 primer (eaeA, bfpB, aggR, astA, pic, hly, stx1, stx2, invE, ipaH, elt, estIa, estIb), and showed the distribution of the strains and its susceptibility to antibiotics. The most frequent pathotypes was Enteropathogenic E.coli 19/42 (45.3%) ) with 9 typical and 10 atypical, followed by Enteroaggregative E. coli 17/42 (40.5%), Enterotoxigenic E. coli 3/42 (7.1%), Enteroinvasive E. coli 3/42 (7.1% ), and 0/42 (0%) in Shigatoxin producing E .coli and no DEC in all control patients. The highest resistance to antibiotics was (95.2%) to Amoxicillin and Ampicillin, respectively, Sulfa-Trimethoprim 92.9%, followed by 85.7% for Tetracycline and Cephalothin, Ceftriaxone 81% and Cefotaxim “clavulanic acid 71.4%. While the lowest resistance was to Chloramphenicol (19 %), Ciprofloxacin (16.7%), Amikacin (7.1%) and no resistance was detected toward Imipenem. We can conclude in this study, multiplex PCR is a swift, and accurate procedure can be used for Diarrheagenic E.coli identification and isolation successfully of strains.
Diarrheagenic E.coli, virulence genes, Multiplex PCR
Diarrheal disease is still a global problem around the world specially in children under five years in developing countries1. According to the World Health Organization (WHO), diarrheal diseases are the second leading cause of death (~760,000 per year) in children 2,3. The microbial causes of diarrhea are variety of bacterial, viral and parasite 4, Among these pathogens, diarrheagenic E. coli play a major role in causing diarrhea in children under 5 years 5,6,7.
When the microbial agent is bacteria, E. coli consider one of the major causes, specially to infantile diarrhea 8,9,10. Depending on specific virulence gene, clinical features, and serotypes diarrheagenic E.coli divided into 6 stains: Enteropathogenic E. coli, Enteroaggregative E. coli, Enteroinvasive E. coli, Enterotoxigenic E. coli, Shiga toxin-producing E. coli and Diffusely Adherence E. coli 11. Culture and biochemical test can’t distinguished between commensal or pathogenic strains of E. coli in stool, therefor PCR used to detect the virulence genes in pathogenic strains 12, multiplex PCR provide detection to many Diarrheagenic E.coli strains virulence genes with high sensitivity, specificity 13.
The aim of this study was detecting the distribution of diarrheagenic E. coli pathotypes among children with diarrhea in Wasit province, Iraq by multiplex PCR, and assessing the antimicrobial susceptibility profile of diarrheagenic E. coli, in order to contribute to the establishment of a more effective empirical antibiotic therapy for the disease.
Collection of samples
During period from middle of September 2017 to middle of December 2017 a total of 210 stool samples were collected from children (males and females) with an ages under five years presented with diarrhea had been admitted at hospitals and attended at private clinics in Wasit province, Iraq. Otherwise, 40 stool samples were collected from healthy children considered as control. The stool samples transported on Carry Blair swabs and cultured on MacConkey agar, XLD, EMB, Blood agar, and CHROMagarSTEC and incubated aerobically at 37 °C for 24 hours, the isolated bacteria was identified according to morphological, biochemical tests and API 20E kit.
Antibiotic susceptibility test: performed by Kirby-Bauer procedure on Muller Hinton agar 14 and results interpreted according to Clinical and Laboratory Standards Institute15
DNA extraction: was performed according to the procedure (Geneaid Genomic DNA extraction Kit).
Multiplex PCR technique: was used for amplifying the genes. The mixture reaction was performed in a total volume 50 µl of PCR Mastermix Gold Multiplex 50x (DNA Template 4 µl, Forward primer 1 µl for each primer, Reverse primer 1 µl for each primer, free water ddH2O 20µl). PCR cycling program parameters used in this reaction for detection of (bfpB , eaeA ,pic , aggR , astA ,invE , ipaH , hly, stx1,stx2, elt ,estla ,estlb ) genes as shown in table (1), the thermal cycling program(Initial denaturation 95°C for 5 min. 1cycle), ( Denaturation 94°C for 30 sec 35 cycle ), (Annealing 58°C for 30 sec. 35 cycle ), ( Extension 72°C for 1 min. 35cycle), (Final extension 72° C for 7min. 1 cycle) (Holding 4° C 1 cycle). The amplification products were electrophoresed through a 2 % agarose gel and visualized with UV transilluminator after ethidium bromide staining. A 100 bp DNA ladder was used as a molecular size marker in gel. The statistical analysis of all the evidence was done using the system SPSS IBM version 20 software, Chi-squire test. P- value £ 0.05 was considered statistically significant.
Table (1):
Primers used for multiplex PCR reaction.
| References | Product size (bp) | Primers ( Sequence (5′ – 3′)) | Primers | E.coli strain |
|---|---|---|---|---|
| 16 | 400 | aggR-F: ACGCAGAGTTGCCTGATAAAG aggR-R: AATACAGAATCGTCAGCATCAGC |
aggR | EAEC |
| 102 | astA-F: TGCCATCAACACAGTATATCCG astA-R: ACGGCTTTGTAGTCCTTCCAT |
astA | ||
| 1,111 | pic-F: AGCCGTTTCCGCAGAAGCC pic-R: AAATGTCAGTGAACCGACGATTGG |
pic | ||
| 17 | 688 | hly-F: TTCTGGGAAACAGTGACGCACATA hly-R: TCACCGATCTTCTCATCCCAATG 0.1 |
hly | STEC |
| 16 | 244 | stx1A-F: CGATGTTACGGTTTGTTACTGTGACAGC stx1A-R: AATGCCACGCTTCCCAGAATTG |
stx1 | |
| 324 | stx2AF: GTTTTGACCATCTTCGTCTGATTATTGAG stx2A-R: AGCGTAAGGCTTCTGCTGTGAC |
stx2 | ||
| 766 | invE-F: CGATAGATGGCGAGAAATTATATCCCG invE-R:CGATCAAGAATCCCTAACAGAAGAATCAC |
invE |
EIEC |
|
| 18 | 437 | ipaH-F: GAAAACCCTCCTGGTCCATCAGG ipaH-R: GCCGGTCAGCCACCCTCTGAGAGTAC |
ipaH | |
|
16 |
482 | eae-F: TCAATGCAGTTCCGTTATCAGTT eae-R: GTAAAGTCCGTTACCCCAACCTG |
eaeA |
EPEC |
| 910 | bfpB-F: GACACCTCATTGCTGAAGTCG bfpB-R: CCAGAACACCTCCGTTATGC |
bfpB | ||
| 655 | elt -F: GAACAGGAGGTTTCTGCGTTAGGTG elt -R: CTTTCAATGGCTTTTTTTTGGGAGTC |
elt | ETEC | |
| 157 | estIa-F:CCTCTTTTAGYCAGACARCTGAATCASTTG estIa -R: CAGGCAGGATTACAACAAAGTTCACAG |
estIa | ||
| 171 | estIb-F: TGTCTTTTTCACCTTTCGCTC estIb-R: CGGTACAAGCAGGATTACAACAC |
estIb |
E. coli were isolated in 100 (47.6 %) of 210 collected samples followed by 78 (37.2%) of other gram negative bacteria (Salmonella, Klebsiella, Proteus, Pseudomonas) and 32 (15.2 %) samples that were no growth. The results of primary diagnosis to these 100 E. coli isolates by selective and differential culture media were consistent with the microscopic and biochemical tests results.
Multiplex applied on theses 100 and 40 control samples and the results showed that DEC in were detected in 42/ 100 (42 %) among diarrheal children compared with 0/100 (0%) among control children. The distribution of 42 DEC pathotype isolates were: EPEC was found in 19 (45.3%), EAEC in 17(40.5% ), ETEC in 3 (7.1%), EIEC in 3 (7.1%) and 0 (0% ) in STEC and controls.
From 19 isolates detected as EPEC which was watery diarrhea 10 (52.6%) isolates of them are atypical EPEC showed eaeA gene found without bfpB gene, and 9 (47.4 %) were typical EPEC which showed eaeA gene together with bfpB gene. All 19 isolates in our study don’t produce nether stx1 or stx2, in addition one of aEPEC showed astA gene.
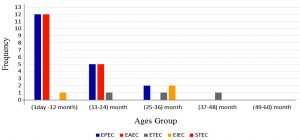
Enteroaggregative E. coli 17 (40.5%) isolates came second after Enteropathogenic E.coli as causative agent of diarrhea among Diarrheagenic E. coli pathotypes in our study, aggR gene was appeared in all EAEC isolates detected in our study that mean all of them were typical EAEC. Enterotoxigenic E.coli account 3 isolates (7.1%) and Enteroinvasive E. coli was detected in 3 isolates (7.1%) that suggested these pathotype maybe play a lessimportant role in childhood diarrhoea in developingcountries. When age stratification was analysed high incidence of DEC E. coli recorded in first and second age group flowed by third and fourth age group, and there were no cases recorded in fifth age group.
The prevalence of Enteropathogenic E. coli infection infections was high in first and second years , also all Enteroaggregative E.coli infections were detected under 2 years, while Enteroinvasive E. coli were high between 2-3 years also cause infection in first age group, in time all Enterotoxigenic E.coli infections were all above 1 year as shown in figure (1).
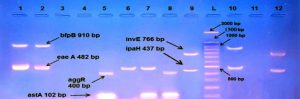
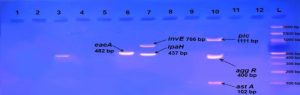
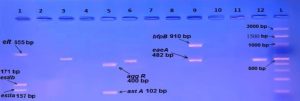
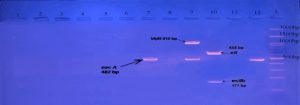
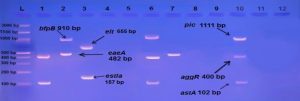
E. coli pathotypes, in our study were identified and isolated successfully by using Multiplex PCR. PCR products visualized to measured product size results from amplification the primers in compared with (100 bp) ladder as shown in figures (2, 3, 4, 5, 6, 7, 8, 9).
Fig. 2. Gel electrophoresis of amplified ( eaeA , bfpB, aggR ,astA ,pic ,hly ,stx1 ,stx2 invE , ipaH, elt ,estIa ,estIb ) genes, the product size (482 , 910 , 400 , 102 , 1,111 , 688 , 244 , 324 , 766 , 437 , 655 , 157 , 171 ( bp ) respectively), of E .coli strains using conventional PCR . Agarose 2%, and TBE (1X ) at (75 V/cm for 90 min., stained with Ethydium bromide dye and visualized on a UV transilluminator. Lane(L): DNA ladder (100-3000 bp), Lanes: (1-12) stool samples.
Fig. 3. Gel electrophoresis of amplified (eaeA, bfpB, aggR, astA, pic, hly, stx1, stx2, invE, ipaH, elt ,estIa, estIb) genes, the product size (482, 910, 400, 102, 1, 111, 688, 244, 324, 766, 437, 655, 157, 171 (bp) respectively), of E .coli strains using conventional PCR. Agarose 2%, and TBE(1X) at (75 V/cm for 90 min., stained with Ethydium bromide dye and visualized on a UV transilluminator. Lane(L): DNA ladder (100-3000bp) Lanes: (1-12 ) stool samples.
Fig. 4. Gel electrophoresis of amplified (eaeA , bfpB, aggR, astA, pic, hly, stx1, stx2, invE, ipaH, elt ,estIa, estIb) genes, the product size (482, 910, 400, 102, 1, 111, 688, 244, 324, 766, 437, 655, 157, 171(bp) respectively), of E .coli strains using conventional PCR. Agarose 2%, and TBE (1X) at (75 V/cm for 90 min., stained with Ethidium bromide dye and visualized on a UV transilluminator. Lane (L): DNA ladder (100-3000bp), Lanes: (1-12) stool samples.
Fig. 5. Gel electrophoresis of amplified (eaeA, bfpB, aggR, astA, pic, hly, stx1, stx2, invE, ipaH, elt, estIa, estIb) genes, the product size (482, 910, 400, 102, 1, 111, 688, 244, 324, 766, 437, 655, 157, 171 (bp) respectively), of E .coli strains using conventional PCR . Agarose 2%, and TBE(1X ) at (75 V/cm for 90 min., stained with Ethidium bromide dye and visualized on a UV transilluminator. Lane (L): DNA ladder (100-3000bp), Lanes: (1-12) stool sample.
Fig. 6. Gel electrophoresis of amplified (eaeA, bfpB, aggR, astA, pic, hly, stx1, stx2, invE, ipaH, elt, estIa, estIb) genes, the product size (482, 910, 400, 102, 1, 111, 688, 244, 324, 766, 437, 655, 157, 171 (bp) respectively), of E .coli strains using conventional PCR. Agarose 2%, and TBE (1X) at (75 V/cm for 90 min., stained with Ethidium bromide dye and visualized on a UV transilluminator. Lane (L): DNA ladder (100-3000 bp), Lanes: (1-12) stool samples.
Fig. 7. Gel electrophoresis of amplified (eaeA, bfpB, aggR, astA, pic, hly, stx1, stx2, invE, ipaH, elt, estIa ,estIb) genes, the product size (482, 910, 400, 102, 1, 111, 688, 244, 324, 766, 437, 655, 157, 171 (bp) respectively), of E. coli strains using conventional PCR . Agarose 2%, and TBE (1X ) at (75 V/cm for 90 min., stained with Ethidium bromide dye and visualized on a UV transilluminator. Lane (L): DNA ladder (100-3000bp), Lanes: (1-12) stool samples.
Fig. 8. Gel electrophoresis of amplified (eaeA, bfpB, aggR, astA, pic, hly, stx1, stx2, invE, ipaH, elt, estIa ,estIb) genes, the product size (482, 910, 400, 102, 1, 111, 688, 244, 324, 766, 437, 655, 157, 171 (bp) respectively), of E. coli strains using conventional PCR. Agarose 2%, and TBE (1X ) at (75 V/cm for 90 min., stained with Ethidium bromide dye and visualized on a UV transilluminator. Lane (L): DNA ladder (100-3000bp), Lanes: (1-12 ) stool samples.
Fig. 9. Gel electrophoresis of amplified (eaeA, bfpB, aggR, astA, pic, hly, stx1, stx2, invE, ipaH, elt, estIa, estIb) genes, the product size (482, 910, 400, 102, 1, 111, 688, 244, 324, 766, 437, 655, 157, 171 ( bp ) respectively ), of E. coli strains using conventional PCR. Agarose 2%, and TBE (1X) at (75 V/cm for 90 min., stained with Ethidium bromide dye and visualized on a UV transilluminator. Lane (L): DNA ladder(100-3000 bp), Lanes: (1-12) stool samples.
Antibiotic susceptibility test results were showed in table (2): The highest level of resistance were to Amoxiclav (95.2%), Ampicillin (95.2%), Sulfa -Trimethoprim (92.9%) followed by Tetracycline (85.7%), Cephalthin (85.7%) Ceftriaxone (81%) Cefotaxime/clavulanicacid 71.4%. The maximum E. coli sensitivity was to Imipenem (100%) flowed by Amikacin (85.7%), Ciprofloxacin (78.6%) Chloramphenicol (73.8%).
Table (2):
Antibiotic susceptibility test.
Antibiotics |
CEC |
AMC |
CTR |
TE |
SXT |
AMP |
CTL |
C |
CIP |
IPM |
AK |
|---|---|---|---|---|---|---|---|---|---|---|---|
Sensitive |
8 19% |
1 2.4% |
7 16.7% |
4 9.5% |
2 4.8% |
2 4.8% |
3 7.1% |
31 73.8% |
33 78.6% |
42 100% |
36 85.7% |
Intermediate |
4 9.5% |
1 2.4% |
1 2.4% |
2 4.8% |
1 2.4% |
0 0% |
3 7.1% |
3 7.1% |
2 4.8% |
0 0% |
3 7.1 % |
Resistance |
30 71.4% |
40 95.2% |
34 81% |
36 85.7% |
39 92.9% |
40 95.2% |
36 85.7% |
8 19% |
7 16.7% |
0 0% |
3 7.1% |
Chi squire |
28 |
72.4 |
44.14 |
52 |
67 |
34.38 |
51.8 |
31.8 |
39.57 |
34.38 |
51.8 |
AK=Amikacin, AMC = Amoxiclav, AMP = Ampicillin, CTR = Ceftriaxone, CTL = Cephalothin,
C = Chloramphenicol, CIP = Ciprofloxacin, CEC = Cefotaxime/Clavulanic, IPM = Imipenem,
TE = Tetracycline, STX = Sulfa-Trimethoprim
The distribution of DEC in our study was 42 (20%) among 210 diarrheal cases. Our result concur to other study in Iraq reported by Hamada et al 12 in Kirkuk (36%) and globally with other studies in Iran Heidary et al 19 (28 %), while our result contrast with other studies were showed less prevalence to Diarrheageic E. coli Konateet al 20 revealed (7.4%) in Burkina Faso, Salmaniet al 21 in Iran who showed (88%). These differences reflecting the difference in distribution of geographical areas , quality of sanitation .
Among all the Diarrheagenic E.coli pathotypes , Enteropathogenic E.coli (EPEC) were found to be the most common pathotypes for children with (45.3%), our result compatible with localstudy by Sakhi 22 who showed EPEC as most than other pathotypes (63%), and in contrast with Khalil 11 and Al-Dulaimi23 where they show it came second after EAEC . Our finding was , however , similar to globally studies with Zhou et al1, Thakur et al7 and Chellapandietal6 that also reported a high frequency of EPEC pathotypes associated with pediatric diarrhea.
EPEC are sub-grouped into typical (tEPEC, eae+ bfpA+) and atypical (aEPEC, eae+ bfpA– ) strains that differ in several respects Naji and Nasser 24 . From 19 isolates detected as EPEC which was watery diarrhea 10 ( 52.6%) isolates of them are atypical EPEC showed eaeA gene found without bfpB gene, and 9 (47.4 %) were typical EPEC which showed eaeA gene together with bfpB gene.
All 19 isolates in our study don’t produce nether stx1 or stx2, in addition one of aEPEC showed astA gene. Our study is close to study by Arif and Salih23 in Sulaimani, Iraq, and global reports by Malvi 25 in India, that showed the distribution of atypical EPEC was higher than typical EPEC. Ochoa and Contreras 26 report that atypical EPEC (aEPEC) are more prevalent than typical-EPEC (tEPEC).
Enteroaggregative E.coli 17 (40.5%) isolates came second after Enteropathogenic E. coli as causative agent of diarrhea among Diarrheagenic E. coli pathotypes in our study, that agree with reports by Sakhi 22 in Dhi-Qar city, also Globally with Thakur et al 7 . But EAEC considered the major cause of diarrhea between diarrheagenic E. coli pathotypes in local studies by Khalil 11 and Al-Dulami 23 , also Globally, Rajendranet et al 27 in India, Ali et al 28 in Egypt, and Konateet et al 20 in Burkina Faso.
EPEC and EAEC were reported as the most common Diarrheagenic E. coli pathotypes Bueriset et al 29; Moyoet et al 30; Wang et al 31.
Enterotoxigenic E. coli account 3 isolates (7.1%) of Diarrheagenic cases, the detection of ETEC is in consonance with previous local findings by Hamada et al 12, and concur with global study by Raghavanet et al 32 in India. Our study differ from other reports by Chiyangiet et al 33 in Zambia which suggested high prevalence of ETEC 40%.
Enteroinvasive E. coli was detected in 3 isolates (7.1%) of Diarrheagenic isolates. Our study is nearly close with local study by Hamada et al 12 (10%), and globally with (0.5%) by Moshtagian 34, and (12.9%) by Konateet et al 20, and (3.7%) by Zhouet et al 1.
Vieira et al 35 also showed low rate of prevalence Enteroinvasive E. coli and suggested that this pathotype may be play a less important role in childhood diarrhea in developing countries. In this study, no Shiga toxin-producing E. coli were detected this result is similar to local reports of Sakhi 22 and Hamada et al 12. Globally, Ali et al 28 and Canzalez-Roman 36, also showed no isolates of STEC were detected in children with diarrhea. STEC appears to be more frequent in adults than children Okekeet et al 37. These difference between our results and other studies may be attributed to rout of infection, virulence factors, pathogen strains, difference in population selection, time of collection and size of samples.
Antibiotic susceptibility test
Resistance to Amoxiclave (95.2% ) agreement with another study had been reported high resistance in studies done by Al-Hilali 38 83.4 % in Al-Najaf. Our result; disagreed with previous study in Alkut by Shamkhi39 who recorded low resistance to Amoxiclav with 7.1% AL-Shuwaikh et al 39 in Baghdad report 33.3%. Increased Amoxiclav resistance coincided with growing Amoxiclav consumption at the community level, similarly, the isolated Diarrheagenic E. coli pathotypes showed high resistance rates to Ampicillin and Sulfa-Trimethoprim Konateet et al 41, as mention in study by Goossenset et al 42; Llor and Bjerrum 43 about antibiotic resistant, there is high resistant in the most consumption antibiotic. Sulfa-Trimethoprim is widely used in developing countries to treat diarrhea because of their availability over the countries Nguyen et al 44. Attention should be given while prescribing Amoxiclav, Ceftriaxone, and Ampicillin to avoid increasing resistance pathotype by E. coli. Similar study conducted by Konate et al 41 in Burkina Faso revealed that 85 % of E. coli isolates were resistance to Tetracycline.
Our study for Cefotaxime was agreement with previous local studies by, AL Hilali 38 68%, Sakhi 22 85.7%, Ugwa et al 45, also reported resistance to Ceftriaxone (91%) by E. coli isolates. Rajeshwari et al 46 reported similar finding for the high resistance of Ceftriaxone 75% and cefotaxime (77.5%) in Indian children with diarrhea, while disagreement Khalil 11 4% and Hamada et al 12 10%. Antibiotic susceptibility testing of isolates showed high resistance rate to Cephalothin (85.7%). The emergence of multidrug resistance especially in E. coli has become a critical public concern, which was designated as resistance to one agent in three or more antibiotic classes. Kamwati 47; Alizadi 48. Many factors responsible for an increase in rates of antimicrobial resistance include misuse/over use of antibiotic by healthcare professionals and general public Magiorakos et al 49; WHO 50; Konateet al 41, and inadequate surveillance systems and independence on reliable microbiological techniques which leads to inappropriate prescription of antibiotics Wellington et al 51.
Ciprofloxacin showed low resistant similar with local studies by Hamada et al 12 10 %, Khalil 11 8%. Globally Kamwati 47 4%, Canizalez-Roman et al 36 21 %. Ciprofloxacin was one of the most active antimicrobial agent which currently recommended to treated diarrhea in children Ayat ollahi et al 52.
Amikacin showed low resistant agreement with 3.3 % reported by Hamada et al12and Al-Hilali 38 0% , and globally with Zhou et al 1 7.4%.
Imipenem with no resistant agreement with 0% resistant from Al-Hilali 38 and Shamkhi 39. The Impenim was the most effective antibiotic against DEC followed by Amikacin, Ciprofloxacin and Chloramphenicol. Impinem has been highly effective against gram negative bacteria Mohammed et al 53, Alam et al 54. They should be used in life threatening multidrug resistance infections where there is no other alternative.
The statistical analysis to susceptibility test results in this study showed high resistance among Diarrheagenic E. coli isolates which were collected from hospitalized children than isolates collected from private pediatric clinics (without history of hospital admitted) as shown in table (3).
Table (3):
Antibiotic sensitivity in hospital and clinical isolates.
No. of resistance isolates from Clinic |
No. of resistance isolates from Hospital |
Antibiotic |
|---|---|---|
0 0% |
2 4.2% |
AK |
0 0% |
0 0% |
IPM |
0 0% |
7 16.7% |
CIP |
2 4.8% |
6 13.4% |
C |
5 11.9% |
31 73.8% |
CTL |
0 0% |
40 95.2% |
AMP |
6 14.3% |
33 78.6% |
SXT |
6 14.3% |
30 71.4% |
TE |
2 4.8% |
32 76.2% |
CTR |
6 14.3% |
34 81% |
AMC |
0 0% |
30 71.4% |
CEC |
This result goes with report of Kamwati47 who showed isolates from children who had been hospitalized were more resistance than those isolated from children not previously hospitalized , and he conclude that recent history of antimicrobial use and hospitalization is a serious predisposing factor to carriage of Multi Drug Resistant strains. Fox-Lewis 55 also mention that hospital-acquired Escherichia coli isolates were multidrug resistant than isolates were community-acquired. Multi drug resistance MDR may be acquired from other patients who have received antibiotics. Infections caused by Multi drug resistance gram negative bacteria are difficult to treat and so may cause more prolonged symptoms in the site of infection Hawkey et al 56.
Enteropathogenic E. coli and Enteroaggregative E. coli was the most common types of Diarrheagenic E. coli among children less than 2 years of age presented with diarrhea in Wasit province. Enterotoxigenic E. coli and Enteroinvasive E. coli were more common in children more than 2 years of age in Wasit province.
This study highlights the Using of multiplex PCR in identifying and successful isolation of Diarrheagenic E.coli from normal flora and can be used as a rapid and accurate method for the isolation of pathogenic strains of E. coli, this will greatly help pediatricians to decrease the use of antibiotic in treatment of diarrhea in children and decreasing the problem of increasing antibiotic resistance. The results of antibiotic sensitivity test revealed that the most active compound against Diarrheagenic E. coli isolates was Imipenem followed by Amikacin, Ciprofloxacin and Chloramphenicol.
Conflict Of Interest
The authors declare that there is no conflict of interest.
- Zhou, Y., Zhu, X., Hou, H., Lu, Y., Yu, J., Mao, L., Mao, L. and Sun, Z. Characteristics of diarrheagenic Escherichia coli among children under 5 years of age with acute diarrhea: a hospital based study. BMC infectious diseases, 2018; 18(1), p.6
- O’Ryan, M., Prado, V. and Pickering, L.K. April. A millennium update on pediatric diarrheal illness in the developing world. In Seminars in pediatric infectious diseases, 2005; 16(2), pp.125-136.
- Kotloff, K.L., Nataro, J.P., Blackwelder, W.C., Nasrin, D., Farag, T.H., Panchalingam, S., Wu, Y., Sow, S.O., Sur, D., Breiman, R.F. and Faruque, A.S. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. The Lancet,2013; 382(9888), pp.209-222.
- Binnicker, M.J. Multiplex molecular panels for the diagnosis of gastrointestinal infection: performance, result interpretation and cost-effectiveness. Journal of clinical microbiology, 2015; pp. 02103.
- Garcia, P.G., Silva, V.L. and Diniz, C.G. Occurrence and antimicrobial drug susceptibility patterns of commensal and diarrheagenic Escherichia coli in fecalmicrobiota from children with and without acute diarrhea. The Journal of Microbiology,2011; 49(1), pp.46-52.
- Chellapandi, K., Dutta, T.K., Sharma, I., Mandal, S., Kumar, N.S. and Ralte, L. Prevalence of multi drug resistant enteropathogenic and enteroinvasive Escherichia coli isolated from children with and without diarrhea in Northeast Indian population. Annals of clinical microbiology and antimicrobials,2017; 16(1), p.49.
- Thakur, N., Jain, S., Changotra, H., Shrivastava, R., Kumar, Y., Grover, N. and Vashistt, J. Molecular characterization of diarrheagenic Escherichia coli pathotypes: Association of virulent genes, serogroups, and antibiotic resistance among moderate to severe diarrhea patients. Journal of clinical laboratory analysis, 2018; p.e22388.
- Benevides-Matos, N., Pieri, F.A., Penatti, M. and Orlandi, P.P. Adherence and virulence genes of Escherichia colifrom children diarrhoea in the Brazilian Amazon. Brazilian Journal of Microbiology,2015; 46(1), pp.131-137 .
- Hoseinzadeh, T., Ghanbarpour, R. and Rokhbakhsh-Zamin, F. Phylogenetic background of enterotoxigenic and enteroinvasive Escherichia coli from patients with diarrhea in Sirjan, Iran. Iranian journal of microbiology,2016; 8(3), p.187.
- Spano, L.C., da Cunha, K.F., Monfardini, M.V., Fonseca, R.D.C.B. and Scaletsky, I.C.A. High prevalence of diarrheagenic Escherichia coli carrying toxin-encoding genes isolated from children and adults in southeastern Brazil. BMC infectious diseases, 2017; 17(1), p.773.
- Khalil, Z.K. Isolation and identification of different diarrheagenic (DEC) Escherichia coli pathotypes from children under five years old in Baghdad. Iraqi Journal Of Community Medicine, 2015; 28(3), pp.126-132.
- Hamada, T.A., Abdelghafoor, A.H. and Hussein, A.S. Evaluation some virulance factor of Eschirichia coli that causes diarrhea in children by using polymerase chain reaction technique in kirkuk city. Medical Journal of Tikrit,2016; 21(1), pp.308-321.
- Arif, S.K. and Salih, L.I. Identification of Different Categories of Diarrheagenic” Escherichia coli” in Stool Samples by Using Multiplex PCR Technique. Asian Journal of Medical Sciences, 2010; 2, pp. 237-243.
- Bauer, A. W., Kirby, W. M. M., Sherris, J. C. and Turck, M. Antibiotic susceptibility testing by a standardized single disc method. Amer. J. Clin. Pathol., 1966; 45: pp.493-496.
- CLSI. Performance standards for antimicrobial susceptibility testing, CLSI Supplement M100S. 2017, 27th ed. Clinical and Laboratory Standards Institute, Wayne, PA, USA.
- Müller, D., Greune, L., Heusipp, G., Karch, H., Fruth, A., Tschäpe, H. and Schmidt, M.A. Identification of unconventional intestinal pathogenic Escherichia coli isolates expressing intermediate virulence factor profiles by using a novel single-step multiplex PCR. Applied and environmental microbiology, 2007; 73(10), pp.3380-3390.
- Antikainen, J., Tarkka, E., Haukka, K., Siitonen, A., Vaara, M., &Kirveskari, J. New 16-plex PCR method for rapid detection of diarrheagenic Escherichia coli directly from stool samples. European journal of clinical microbiology & infectious diseases, 2009; 28(8), 899-908.þ
- Vidal, M., Kruger, E., Durán, C., Lagos, R., Levine, M., Prado, V., Toro, C. and Vidal, R. Single multiplex PCR assay to identify simultaneously the six categories of diarrheagenic Escherichia coli associated with enteric infections. Journal of clinical microbiology, 2005; 43(10), pp.5362-5365.
- Heidary, M., Momtaz, H. and Madani, M. Characterization of diarrheagenic antimicrobial resistant Escherichia coli isolated from pediatric patients in Tehran, Iran. Iranian Red Crescent Medical Journal,2014; 16(4).
- Konaté, A., Dembélé, R., Kagambèga, A., Soulama, I., Kaboré, W.A., Sampo, E., Cissé, H., Sanou, A., Serme, S., Zongo, S. and Zongo, C. Molecular characterization of diarrheagenic Escherichia coli in children less than 5 years of age with diarrhea in Ouagadougou, Burkina Faso. European Journal of Microbiology and Immunology, 2017; 7(3), pp.220-228.
- Salmani, H., Azarnezhad, A., Fayazi, M.R. and Hosseini, A. Pathotypic and phylogenetic study of diarrheagenic Escherichia coli and uropathogenic E. coli using multiplex polymerase chain reaction. Jundishapur journal of microbiology, 2016; 9(2).
- Sakhi R. J. Isolation of Escherichia coli from diarrhea and test their pathogenicity and susceptibility pattern for antibiotic, International Journal of Agricultural science and research, 2016; 6(2), p. 29 – 34.
- Al-Dulaimi, T.H., Aziz, H.W., Al-Marzoqi, A.H., Al-Aziz, S.A. and Mohsin, S.A.A., Molecular characterization and antibiotic susceptibility of diarrheagenic Escherichia coli from Children. Medical Journal of Babylon,2015; 12(2), pp.541-550.
- Naji H. F., Nasser . A. S. Molecular Identification of Specific Virulence Genes in Enteropathogenic Escherichia coli. IOSR Journal of Pharmacy and Biological Sciences, 2015; 10, pp.67-73.
- Malvi, S., Appannanavar, S., Mohan, B., Kaur, H., Gautam, N., Bharti, B., Kumar, Y. and Taneja, N. Comparative analysis of virulence determinants, antibiotic susceptibility patterns and serogrouping of atypical enteropathogenic Escherichia coli versus typical enteropathogenic E. coli in India. Journal of medical microbiology, 2015; 64(10), pp.1208-1215.
- Ochoa, T. J. and Contreras, C. A. Enteropathogenic E. coli (EPEC) infection in children. Current opinion in infectious diseases, 2011; 24(5),p. 478.þ
- Rajendran, P., Ajjampur, S.S.R., Chidambaram, D., Chandrabose, G., Thangaraj, B., Sarkar, R., Samuel, P., Rajan, D.P. and Kang, G. Pathotypes of diarrheagenic Escherichia coli in children attending a tertiary care hospital in South India. Diagnostic microbiology and infectious disease,2010; 68(2), pp.117-122.
- Ali, M.M.M., Ahmed, S.F., Klena, J.D., Mohamed, Z.K., Moussa, T.A. and Ghenghesh, K.S. Enteroaggregative Escherichia coli in diarrheic children in Egypt: molecular characterization and antimicrobial susceptibility. The Journal of Infection in Developing Countries,2014; 8(05), pp.589-596.
- Bueris, V., Sircili, M.P., Taddei, C.R., Santos, M.F.D., Franzolin, M.R., Martinez, M.B., Ferrer, S.R., Barreto, M.L. and Trabulsi, L.R. Detection of diarrheagenic Escherichia coli from children with and without diarrhea in Salvador, Bahia, Brazil. Memórias do InstitutoOswaldo Cruz,2007; 102(7), pp.839-844.
- Moyo, S.J., Maselle, S.Y., Matee, M.I., Langeland, N. and Mylvaganam, H. Identification of diarrheagenic Escherichia coli isolated from infants and children in Dar es Salaam, Tanzania. BMC infectious diseases, 2007; 7(1), p.92.
- Wang, X., Wang, J., Sun, H., Xia, S., Duan, R., Liang, J., Xiao, Y., Qiu, H., Shan,G. and Jing, H., Etiology of childhood infectious diarrhea in a developed region of China: compared to childhood diarrhea in a developing region and adult diarrhea in a developed region. PLoS One, 2015; 10(11).
- Raghavan, P.R., Roy, S., Thamizhmani, R. and Sugunan, A.P. Diarrheagenic Escherichia coli infections among the children of Andaman Islands with special reference to pathotype distribution and clinical profile. Journal of epidemiology and global health, 2017; 7(4), pp.305-308.
- Chiyangi, H., Muma, J. B., Malama, S., Manyahi, J., Abade, A., Kwenda, G. and Matee, M. I. Identification and antimicrobial resistance patterns of bacterial enteropathogens from children aged 0–59 months at the University Teaching Hospital, Lusaka, Zambia: a prospective cross sectional study. BMC infectious diseases,2017; 17(1), pp.117.
- Moshtagian, F., Alipour, M., and Yahyapour, Y., Prevalence of Escherichia coli Pathotypes Among Children With Diarrhea in Babol, Northern Iran. Int J Enteric Pathog, 2016; 4(3),p 1-4.
- Vieira, N., Bates, S. J., Solberg, O. D., Ponce, K., Howsmon, R., Cevallos, W., Trueba, G., Riley, L. and Eisenberg, J. N. High prevalence of enteroinvasive Escherichia coli isolated in a remote region of northern coastal Ecuador. The American journal of tropical medicine and hygiene, 2007; 76(3), 528-533.
- Canizalez-Roman, A., Flores-Villaseñor, H.M., Gonzalez-Nuñez, E., Velazquez-Roman, J., Vidal, J.E., Muro-Amador, S., Alapizco-Castro, G., Díaz-Quiñonez, J.A. and León-Sicairos, N. Surveillance of Diarrheagenic Escherichia Coli strains isolated from diarrhea cases from children, adults and elderly at northwest of Mexico. Frontiers in microbiology,2016; 7, p. (1924).
- Okeke, I.N., Ojo, O., Lamikanra, A. and Kaper, J.B. Etiology of acute diarrhea in adults in southwestern Nigeria. Journal of clinical microbiology, 2003; 41(10), pp.4525-4530.
- Al Hilali, S.A. Occurrence and molecular characterization of enteropathogenic Escherichia coli serotypes isolated from children with diarrhoea in Najaf, Iraq, 2010. M. SC. Thesis, Kufa University, College of Medicine.
- Shamki, J.A., Al-Charrakh, A.H. and Al-Khafaji, J.K. Detection of ESBLs in Enteropathogenic E. coli (EPEC) isolates associated with infantile diarrhea in Kut City. Medical Journal of Babylon,2012; 9(2), pp.403-412.
- AL-Shuwaikh, A. M., Ibrahim, I. A. and Al-Shwaikh, R. M. Detection of E. coli and Rotavirus in Diarrhea among Children Under Five Years Old. Iraqi Journal of Biotechnology, 2015; 14(1), 85-92.
- Konaté, A., Dembélé, R., Guessennd, N.K., Kouadio, F.K., Kouadio, I.K., Ouattara, M.B., Kaboré, W.A., Kagambèga, A., Cissé, H., Ibrahim, H.B. and Bagré, T.S. Epidemiology and antibiotic resistance phenotypes of diarrheagenic Escherichia coli responsible for infantile gastroenteritis in Ouagadougou, Burkina Faso. European Journal of Microbiology and Immunology, 2017; 7(3), pp.168-175.
- Goossens, H., Ferech, M., Vander Stichele, R., Elseviers, M. and ESAC Project Group. Outpatient antibiotic use in Europe and association with resistance: a cross-national database study. The Lancet,2005; 365, pp.579-587.þ
- Llor, C., and Bjerrum, L. Antimicrobial resistance: risk associated with antibiotic overuse and initiatives to reduce the problem. Therapeutic advances in drug safety, 2014; 5(6), pp229-24.
- Nguyen, T.V., Van Le, P., Le, C.H. and Weintraub, A., Antibiotic resistance in diarrheagenic Escherichia coli and Shigella strains isolated from children in Hanoi, Vietnam. Antimicrobial Agents and Chemotherapy,2005; 49(2), pp.816-819.
- Ugwu, M. C., Edeani, G. I., Ejikeugwu, C. P., Okezie, U., and Ejiofor, S. O., Antibiotic Susceptibility Profile of Escherichia coli and Salmonella Causing Childhood Diarrhoea in Awka Municipality, South-eastern Nigeria. ClinMicrobiol, 2017; 6(277),p 2.
- Rajeshwari, K., Beena, U., Singh, R., Tiwari, G. and Ajay, K. M. Multi drug resistant enteropathogenic E. coli diarrhea in children. Am J Res Commun, 2015; 3(9),pp 27-48.
- Kamwati K. S. Antimicrobial resistant Escherichia coli genes in children age below five years presenting with diarrhoea at thika level 5 hospital, Kiambu country, phDthesis, 2015.Kenya
- Alizade, H. Escherichia coli in Iran: An Overview of Antibiotic Resistance: A Review Article. Iranian journal of public health,2018; 47(1), p.1.
- Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., Harbarth, S., Hindler, J.F., Kahlmeter, G., Olsson-Liljequist, B., Paterson, D.L., Rice, L.B., Stelling, J., Struelens, M.J., Vatopoulos, A., Weber, J.T. and Monnet D.L. Multidrug resistant, extensively drug resistant and pandrug resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clinical microbiology and infection, 2012; 18(3), pp.268-281.
- World Health Organization 2014. Antimicrobial resistance: global report on surveillance.Geneva, Switzerland. Printed in France
- Wellington, E.M., Boxall, A.B., Cross, P., Feil, E.J., Gaze, W.H., Hawkey, P.M., Johnson-Rollings, A.S., Jones, D.L., Lee, N.M., Otten, W. and Thomas, C.M. The role of the natural environment in the emergence of antibiotic resistance in Gram-negative bacteria. The Lancet infectious diseases, 2013; 13(2), pp.155-165.
- Ayatollahi, J., Shahcheraghi, S. H., Akhondi, R. and Soluti, S. S. Antibiotic resistance patterns of Escherichia coli isolated from children in shahidsadoughi hospital of Yazd. Iranian journal of pediatrichematology and oncology,2013; 3(2), pp.78.
- Mohammed, M.A., Alnour, T.M., Shakurfo, O.M. and Aburass, M.M. Prevalence and antimicrobial resistance pattern of bacterial strains isolated from patients with urinary tract infection in Messalata Central Hospital, Libya. Asian Pacific journal of tropical medicine, 2016; 9(8), pp.771-776.
- Alam, M.Z., Alam, Q., Jiman-Fatani, A.A., Shukri, H.A., Haque, A.A. Surveillance study on the prevalence and antimicrobial resistance pattern among different groups of bacteria isolated from Western province of Saudi Arabia. Biomed. Res. 2017; 28(2):pp898-906.
- Fox-Lewis, A., Takata, J., Miliya, T., Lubell, Y., Soeng, S., Sar, P., Rith, K., McKellar, G., Wuthiekanun, V., McGonagle, E. and Stoesser, N. Antimicrobial resistance in invasive bacterial infections in hospitalized children, Cambodia, 2007–2016. Emerging infectious diseases, 2018; 24(5), p.841.
- Hawkey, P.M., Warren, R.E., Livermore, D.M., McNulty, C.A., Enoch, D.A., Otter, J.A. and Wilson, A.P.R. Treatment of infections caused by multidrug-resistant Gram-negative bacteria: report of the British Society for Antimicrobial Chemotherapy/healthcare Infection Society/british Infection Association Joint Working Party. Journal of Antimicrobial Chemotherapy, 2018; 73(suppl_3), pp.iii2-iii78.
© The Author(s) 2018. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.