ISSN: 0973-7510
E-ISSN: 2581-690X
Viral outbreaks can result from the consumption of contaminated bivalve mollusks. However, despite the regulation related to enteric bacteria in food products, the consumption of raw and undercooked mollusks remains linked to viral epidemics in human populations. Real-time RT-PCR is a highly sensitive approach for detecting and quantifying enteric viruses, and after eliminating enzymatic amplification inhibitors from samples of interest, sensitive and specific tests, like real-time RT-PCR, can facilitate the detection and quantification of a wide range of viruses that are concentrated in mollusk digestive tissues. The aim of the present study was to evaluate the prevalence of Group-A rotaviruses in mussel (Mytilus edulis Linnaeus, 1758) specimens (n=576) collected downstream of the Oued El Maleh Estuary, which is along the coast of Mohammedia City in Morocco, using real-time RT-PCR. Rotavirus A RNA was detected in 37.5% (n=18) of the 48 sample batches, and viral loads ranged from 0.42×101 to 1.8603×104 genomic copies per g digestive tissue. Most (72.22%) of the positive samples were collected during the wet season (September-April), and the probability of detecting rotaviruses was significantly greater during the wet season than during the dry season (P<0.001). Monitoring Rotavirus A and similar viruses in shellfish may help prevent viral contamination and preserve public health.
Rotavirus, Molluscs, Gastroenteritis, Environment, Real-Time RT-PCR
Rotaviruses (Rotavirus, Reoviridae) are non-enveloped viruses that each consist of 11 double-stranded RNA molecules.1 Group-A rotaviruses are the main causative agents of acute gastroenteritis (GEA) in children and are responsible for 1 to 3 million deaths each year, including more than 800,000 in Africa,2 and almost 220,000 hospitalizations per year in industrialized countries.3 The National Rotavirus Surveillance Network of the Ministry of Health in Morocco estimated that, between 2006 and 2010, 48,871 cases of diarrhea in children under 5 years of age were caused by rotaviruses each year.4 With gradual improvements in diarrhea management, including oral rehydration, mortality rates decreased in Morocco from 528,000 in 2000 to 215,000 in 2013.5 Because the negative effects of rotavirus infection can be exacerbated by malnutrition, mortality rates tend to be highest in developing countries.6
Two rotavirus vaccines (RotaTeq from Merck and Rotarix from GSK Biologicals) are currently approved for global use by the WHO and have exhibited efficacy in many countries.7-8 However, both vaccines have also demonstrated significantly lower immunogenicity and efficacy in low-income countries in Africa and Asia.9
Rotavirus transmission is influenced by multiple factors, including the survival and persistence of the virus on food, attachment of the virus to food, mode of contamination, mode of food consumption (e.g., raw, cooked, peeled), and the consumer of the contaminated food.10 Water-borne Rotavirus A-related diseases have been reported.11 Coastal discharges are constantly releasing human viruses, which can survive for weeks or even months in the environment.12 Therefore, the identification of circulating virus strains can be achieved by environmental monitoring.
The Food and Agriculture Organization of the United Nations (FAO) reported that, in 2015, 15 million tonnes of bivalve mollusks (>104 species) were produced, together accounting for 21% of global aquaculture production intended for human consumption.13 Accordingly, this sector is of great economic interest. However, such mollusks and crustaceans are important vehicles for the transmission of foodborne viruses,14 especially when considering that their filtering activities can result in viral loads that are 100 to 1000 times greater than that of the surrounding water.15 Shellfish grown in polluted waters can accumulate a variety of microbes, including pathogenic bacteria and viruses that can compromise consumer health.16 Indeed, infectious agents carried by humans, in asymptomatic case or in the incubation phase, are sheded in large quantities in the environment, thus threatening the health of young children when they are in direct contact.17 These pathogens can also harm aquatic ecosystems, thereby affecting bivalve growth and production.18,19 In polluted ecosystems, bivalve mollusks filter fecal pathogens along with organic particles.20,21
Bivalve mollusks are valuable bio-indicators of microbial pollution in aquatic environments, because the direct detection in water is only occasional. Even though monitoring bacterial indicators of fecal contamination in shellfish and their habitats can help prevent bacterial gastroenteritis, it remains difficult to monitor contamination by enteric viruses.22 The present study focuses on mussels, owing to their wide geographical distribution and filtering behaviors, which make them valuable bio-indicators.23
The objective of the present study was to develop methods for identifying fecal contamination by Rotavirus A in mussels collected from a potentially polluted area and the development of a national database of the results of virological analyzes of the coastal environment.
Study site and sample collection
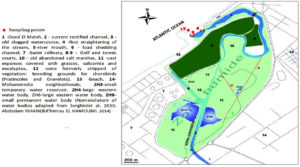
From July 2018 to July 2019 (13 months), mussel (Mytilus edulis Linnaeus, 175) samples were collected from three wild-population sites (S1: 33°42’16.7″N, 7°24’43.3″W; S2: 33°42’13.4″N, 7°24’47.8″W; S3: 33°42’09.1″N, 7°24’55.0″W) at the mouth of the Oued El Maleh River (i.e., Oued El Maleh Estuary; Fig. 1), which receives untreated domestic and industrial effluents and is known as an important location for harvesting mollusks intended for human consumption. The Oued El Maleh Estuary was designated as a “Ramsar Site” in 2005 under the number 1486.24 It is in an urban environment (Fig. 1) that includes a complex of ephemeral rain-fed water bodies. This Ramsar site, a large part of which is privately owned, is subject to intense anthropozoogenic activity, which includes grazing, poaching, and the illegal dumping of low-degradable waste (e.g., tires, metal waste, and plastics) and liquid products. Despite its unsanitary nature, the site remains an important location for harvesting mollusks intended for human consumption.
Immediately after collection, the mussels (n=576; 48 batches of 12) were refrigerated and shipped directly to the laboratory, and processed within 24 h. During processing, the mollusks were cleaned, using brushes and tap water, to remove foreign bodies from their shells, thoroughly washed using sterile water, and dissected to extract hepatopancreas samples. After extraction, batches of hepatopancreas samples (1.5 g, corresponding to samples from 10–12 mussels) were frozen at -20°C until further analysis.
Standard precautions were used during all manipulations to reduce the probability of sample contamination, and separate areas of the lab were used for reagent preparation, sample processing, and nucleic acid analysis.
Because loads of human enteric viruses in bivalves are so low, effective virus detection must be preceded by the elimination of naturally occurring PCR inhibitors and by virus concentration.25 This was achieved by direct elution into glycine buffer and acid adsorption, followed by elution, which aims to separate virions and flesh as much as possible, followed by polyethylene glycol (PEG) concentration.26
Virus concentration, RNA extraction and RT-PCR
Virus was extracted from the samples using a previously described procedure,26 with modifications. The hepatopancreas batch samples (1.5 ± 0.2 g) were homogenized, diluted in equal volumes of glycine buffer (0.05 M glycine, 0.15 M NaCl, pH 9.0), and shaken for 15 min at 4°C on a gyratory shaker to release virus from tissue. The homogenates were then centrifuged at 5000 ×g for 15 min, and corresponding supernatants were collected. For further concentration of the virus particles, each supernatant (2 ml) was mixed with 5 ml phosphate-buffered saline (PBS, pH 7.2) and 50% PEG 6000, which was added to the final volume in a 1:3 proportion, and the resulting solutions were adjusted to a pH of 7.2 and incubated overnight at 4°C. The following day, the mixtures were centrifuged for 30 min at 10,000 ×g and 4°C, and the resulting pellet or precipitate was suspended in 500 µl PBS (pH 7.2) and then stored at -20°C for subsequent analysis.
Total RNA was extracted from the virus concentrate using Trizol Invitrogen™, according to the manufacturer’s instructions, and the qualities of the extracted RNAs were evaluated by spectrophotometry at 260 and 280 nm using a NanoDrop 2000/200c spectrophotometer (Thermo Fisher Scientific, USA).
RT-PCR molecular analysis was performed using the Primerdesign genesig Advanced Kit for Human Rotavirus A, which targets the NSP5 gene. The 20-µl reactions included 5 µl sample RNA (100×–1000× dilution, depending on sample concentration) and 15 µl master mix and were amplified using the SaCycler-96 (Sacace Biotechnologies), following the manufacturer’s instructions: reverse transcription at 55°C for 10 min, enzyme activation at 95°C for 2 min, 50 denaturing cycles at 95°C for 10 s, and then FAM and VIC quantification at 60°C for 60 s. All samples were analyzed in duplicate, and two negative controls (RNase free water) were included, as well as five standard concentrations (2 to 2×104 copies/µl) of a positive control (2×105 copies/µl). cDNA was synthesized from the RNA of positive samples using the Precision nanoScript 2 Reverse Transcription kit, according to the manufacturer’s instructions, and a Perkin Elmer 9600 thermocycler. The synthesized cDNA was stored at -20°C for subsequent sequencing.
Artificial shellfish contamination
To evaluate the sensitivity Mussels (Mytilus edulis) were purchased from a local supermarket sourced from approved shellfish producers, tested using the same kit, and confirmed negative for Rotavirus A. The digestive glands of the mussels were extracted, inoculated with 100 μl of the same series 10 times diluted from a stool sample containing a known viral load of a strain classified as G1P[8]. The stool sample was graciously provided by Professor Jamal Eddine Hafid (Bioresources Laboratory and Food Safety, Faculty of Sciences and Techniques-Gueliz, University CADI AYYAD Marrakech, Morocco).
Evaluation of extraction efficiency and detection limits
All the viral dilution suspensions were divided into 100-µl single-use aliquots, since repeated freezing/thawing can reduce viral content by 50–80% (data not shown).
It is possible to assess extraction efficiency by adding a known quantity of Mengovirus to each sample before processing and comparing real-time RT-PCR amplification of the spiked sample to that of an equal quantity of purified Mengovirus. Accordingly, 10 μl Mengovirus (Mengovirus@ceeramTools™ Kit (Ceeram, France)) was added to 1.5 ± 0.2 g artificially contaminated digestive tissues before extraction, and a 10-fold serial dilution of Mengovirus was used to establish a standard curve. The standard curve was then used to estimate the Ct value of Mengovirus RNA extracted from artificially contaminated samples and, thus, assess extraction efficiency. Extraction yields of >1% were considered valid. Detection limits for real-time PCR tests and dilution accuracy results were evaluated by performing a 1/10 dilution series of the positive control, it corresponds to 101 copies of RNA corresponding to Ct = 33.3 (Table 2).
Identification of false negatives
To assess the frequency of false-negative PCR results resulting from the presence of inhibitors, 1 µl titrated internal control (IC) was added to all real-time RT-PCR reactions.
Statistical Analysis
The experimental data were tabulated and analyzed by One-way ANOVA, using the SPSS version 17.0, to evaluate the effects of rainfall on the number of contaminated samples. P-values below 0.05 were considered significant.
The simultaneous extraction of rotavirus and Mengovirus provided a mean yield-to-yield ratio (Mengo/Rota) of 0.063, which allowed the subsequent estimation of rotavirus extraction yield.
The recovery rates of rotavirus and Mengovirus from mussel (Mytilus edulis) extracts were estimated to range from 19 to 31% and from 1 to 2.5%. The RT-PCR detection limit was up to 1.75 PFU/g digestive tissue.
Table (1):
Estimated prevalence of rotavirus in mussels in the Oued El Maleh estuary.
Total Number of sub sample (1.5g) |
Positive sub sample (1.5g) |
Prevalence Estimation (%) |
|
|---|---|---|---|
RNA (RVA) |
48 |
18 |
37,5 |
Rotavirus prevalence and quantity
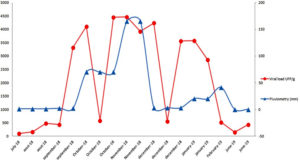
The evaluation of real-time PCR detection limits, which was performed using 10-fold dilutions of positive control, revealed a sensitivity of 10 genomic copies per reaction (Table 1), corresponding to 33.3 Ct. Therefore, 18 (37.5%) of the 48 sample batches were positive for Rotavirus A, with viral loads ranging from 0.421×101 to 1.8603×104 PFU/g digestive tissue (Table 2). In terms of seasonal variation, Rotavirus A was significantly less prevalent during the dry season (May–August; 5/26, 19.23%) than during the wet season (September–April; 13/21, 61.90%; P<0.001). Therefore, there was a significant correlation between mean rainfall and Human rotavirus A prevalence in the Oued El Maleh Estuary during the study period (Fig. 2).
Table (2):
Detection limits of real-time PCR tests and within-test dilution accuracy results of 10 times positive control tested in three specimens.
Nb of RNA copies per reaction |
detected samples |
average Ct |
|---|---|---|
104 |
3/3 |
24.8 |
103 |
2/3 |
27.6 |
102 |
2/3 |
30.8 |
101 |
1/3 |
33.3 |
1 |
1/3 |
37.16 |
In Morocco, safety parameters for the production of shellfish intended for human consumption have not yet been extended to human enteric viruses. These safety parameters must be chosen among those which can be carried out in the laboratory as part of the routine follow-up of environmental fecal contamination. This monitoring aims to carry out upstream actions at the level of wastewater treatment lines in order to ensure the prevention of potential viral etiology epidemics.
Table (3):
Results of RVA quantification in positive samples during the study period.
Week |
Load in Reaction Volume (cp/ µl) |
CT |
Viral Load PFU/g |
|---|---|---|---|
1S7-18 |
0,396 |
35.0 >33.3 |
– |
2S7-18 |
– |
– |
– |
3S7-18 |
0,998 |
33.6 >33.3 |
– |
4S7-18 |
1,2 |
33.3 |
421 |
5S7-18 |
0,96 |
33.6 >33.3 |
– |
1S8-18 |
– |
– |
– |
2S8-18 |
1,9 |
32.6 |
667 |
3S8-18 |
5,7 |
30.9 |
2000 |
4S8-18 |
– |
– |
– |
1S9-18 |
5,1 |
31.1 |
1790 |
3S9-18 |
39,4 |
27.9 |
13829 |
1S10-18 |
– |
– |
– |
2S10-18 |
50,3 |
27.6 |
17655 |
3S10-18 |
6,9 |
30.6 |
2422 |
4S10-18 |
53 |
27.5 |
18603 |
1S11-18 |
– |
– |
– |
2S11-18 |
48,7 |
27.6 |
17094 |
3S11-18 |
46,6 |
27.7 |
16356 |
4S11-18 |
– |
– |
– |
1S12-18 |
48,7 |
27.6 |
17093 |
2S12-18 |
6,6 |
30.7 |
2316 |
3S12-18 |
42,3 |
27.8 |
14847 |
4S12-18 |
– |
– |
– |
1S01-19 |
42,3 |
27.8 |
14847 |
2S01-19 |
– |
– |
– |
3S01-19 |
33,9 |
28.2 |
11899 |
1S02-19 |
6,2 |
30.8 |
2176 |
2S02-19 |
– |
– |
– |
3S02-19 |
– |
– |
– |
4S02-19 |
– |
– |
– |
2S03-19 |
– |
– |
– |
3S03-19 |
0,372 |
35.1 > 33.3 |
|
1S04-19 |
– |
– |
– |
2S04-19 |
– |
– |
– |
3S04-19 |
– |
– |
– |
4S04-19 |
– |
– |
– |
1S05-19 |
– |
– |
– |
2S05-19 |
0,372 |
35.1 >33.3 |
|
3S05-19 |
– |
– |
– |
4S05-19 |
– |
– |
– |
1S06-19 |
1,8 |
32.7 |
632 |
2S06-19 |
– |
– |
– |
3S06-19 |
– |
– |
– |
4S06-19 |
0,213 |
35.9 >33.3 |
|
1S07-19 |
5,1 |
31.2 |
1790 |
2S07-19 |
– |
– |
– |
3S07-19 |
– |
– |
– |
4S07-19 |
– |
– |
– |
In the present study, the viral contamination rate of the mussel samples was relatively high (37.5%), and even though the observed rate of prevalence was not as high as that reported by a follow-up of mussels in France,27 similar results have been reported by a variety of similar studies.23,27,28 Sedentary mollusks are the most affected by aquatic pollution in comparison to mobile forms, and viral loads are usually greater in mollusks living in sediments than in mussels. Indeed, burrowing species concentrate significantly higher amounts of virus than species that live in open water.29 In aquatic environments, bacteria and viruses can either float freely or adsorb to the surfaces of suspended particles,30 and such particles, along with other plankton, are filtered by bivalve mollusks and accumulated in their digestive tissues. Santiago et al.31 used RT-PCR to characterize the Rotavirus A loads of oysters and reported a much higher contamination rate (10/30, 33%) than Kou et al.,32 who reported that only 21% (32/150) of oyster samples were contaminated by rotavirus and norovirus, with 3.3% (5/150) contaminated by rotavirus, 4% (6/150) contaminated by GI norovirus, and 14% (21/150) contaminated by GII norovirus. The present study confirms the contamination of bivalves in the estuary by rotavirus. The minimum infectious dose of rotaviruses is very low, and theoretically a single infectious particle would be sufficient to trigger in vivo viral multiplication and/or seroconversion.33-35 In fact, even a single viral particle could be sufficient to trigger disease.36 It is, therefore, very likely that viral particles that can persist in outdoor environments can contaminate humans through the consumption of contaminated food. Studies of the physico-chemical properties and resilience of rotaviruses in outdoor environments have confirmed the contribution of rotaviruses to nosocomial diseases.37 Naturally contaminated shellfish have been reported to harbor viral loads ranging from 102 to 104 genomic copies per gram digestive tissue.38-41
The results of the present study confirm the unsanitary status of the sampling area near the Oued El Maleh Estuary in Mohammedia, Morocco, as well as the contribution of mollusks as vectors of rotavirus strains.
Morocco is a temperate country, and winter peaks in outbreaks are related to enteric pollution.42 The seasonality of environmental contamination with Rotavirus A was similar to that reported by Benhafid et al.43
The Oued El Maleh Estuary is a gathering place for migratory birds and is located 1 km from an old landfill, from which leachate is transported by the river. Additional sources of pollution include waste water from people not served by sanitation, butcher companies, and the municipal slaughterhouse. Rotavirus A was selected as the focus of the present study because it is the most common cause of diarrhea in humans and for its high excretion rate (~100 billion virions per gram feces).44 The analysis of bivalve mollusks can provide an overview of contamination in the studied environment.45
Because the collection sites used by the present study were located in the Oued El Maleh Estuary, the area’s contamination could also be derived from upstream areas. Therefore, upstream rainfall could affect the environmental conditions of the estuary.
The molecular isolation of rotaviruses from shellfish remains difficult to validate since there is no standard method for virus extraction. However, rotavirus detection kits are specific to the virological study of the environment and are generally fast, sensitive, and capable of detecting non-cultivable viruses. The results of the present study demonstrate that RT-PCR can be used to characterize and quantify rotavirus contamination in shellfish, provide insight into the circulation of rotaviruses in the environment, suggest the RT-PCR could be used as a reliable method for water quality monitoring of Moroccan coastal against any threat of poor treatment of domestic waste and, therefore, the preservation of the health of Moroccan consumers.
The present study provides a comprehensive assessment of rotavirus contamination in wild mussels that are locally available to consumers in Mohammadia. By evaluating rotavirus contamination over 13 consecutive months, the study was able to identify seasonal variation in contamination rate. Rotavirus A contamination was significantly more prevalent during the wet season than during the dry season (P<0.005). Monitoring fecal viruses in shellfish could help prevent viral contamination and preserve public health. However, shellfish from uncontrolled areas must be systematically prohibited for human consumption due to the high risk of viral contamination.
ACKNOWLEDGMENTS
We would like to thank Faculty of Science and Technology of Mohammedia, where this research was carried out with its financial support; we would like to thank the Director of the Institute Pasteur of Morocco for agreeing to collaborate with our research team.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
- Estes MK, Kapikian AZ, Knipe DM, Howley PM. Rotaviruses. Fields Virology. Eds) Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE, Philadelphia: Kluwer Health/Lippincott, Williams and Wilkins, 2007; 1917-1974.
- Black RE, Cousens S, Johnson HL, et al. Global, regional, and national causes of child mortality in 2008: a systematic analysis. The Lancet. 2010;375(9730):1969-1987.
Crossref - Grimwood K, Bines JE. Rotavirus vaccines must perform in low-income countries too. The Lancet. 2007;370(9601):1739-1740.
Crossref - Benhafid M, Rguig A, Trivedi T, et al. Monitoring of rotavirus vaccination in Morocco: Establishing the baseline burden of rotavirus disease. Vaccine. 2012;30(46):6515-6520.
Crossref - Tate JE, Burton AH, Boschi-Pinto C, et al. Global, Regional, and National Estimates of Rotavirus Mortality in Children <5 Years of Age, 2000-2013. Clin Infect Dis. 2016;62:S96-S105.
Crossref - Iturriza-Gomara M, Kang G, Gray J. Rotavirus genotyping: keeping up with an evolving population of human rotaviruses. J Clin Virol. 2004;31(4):259-265.
Crossref - Armah GE, Sow SO, Breiman RF, et al. Efficacy of pentavalent rotavirus vaccine against severe rotavirus gastroenteritis in infants in developing countries in sub-Saharan Africa: a randomized, double-blind, placebo-controlled trial. The Lancet. 2010;376(9741):606-614.
Crossref - Zaman K, Dang DA, Victor JC, et al. Efficacy of pentavalent rotavirus vaccine against severe rotavirus gastroenteritis in infants in developing countries in Asia: a randomised, double-blind, placebo-controlled trial. The Lancet. 2010;376(9741):615-623.
Crossref - Fischer Walker CLF, Black RE. Rotavirus vaccine and diarrhea mortality: quantifying regional variation in effect size. BMC Public Health. 2011;11(3):S16.
Crossref - Le Guyader FS, Atmar RL. Binding and inactivation of viruses on and in food, with a focus on the role of the matrix. In: Koopmans MPG, Cliver DO, Bosch A, editors. “Food-borne viruss: progress and challenges”. ASM Press. 2009:189-208.
Crossref - Divizia M, Gabrieli R, Donia D, et al. Water borne gastroenteritis outbreak in Albania. Wat Sci Technol. 2004;50(1):57-61.
Crossref - Gantzer C, Dubois E, Crance JM, et al. Influence of environmental factors on the survival of enteric viruses in seawater. Oceanol Acta. 1998;21(6):983-992.
Crossref - Subasinghe R. World aquaculture 2015: a brief overview. FAO Fisheries and Aquaculture Report, 2017(1140). https://www.proquest.com/docview/1931154017?pq-origsite=gscholar&fromopenview=true
- Seo DJ, Lee MH, Son NR, et al. Seasonal and regional prevalence of norovirus, hepatitis A virus, hepatitis E virus, and rotavirus in shellfish harvested from South Korea. Food Control. 2014;41:178-184.
Crossref - Rodriguez-Lazaro D, Cook N, Ruggeri FM, et al. Virus hazards from food, water and other contaminated environments. FEMS Microbiol Rev. 2012;36(4):786-814
Crossref - Muniain-Mujika I, Girones R, Lucena F. Viral contamination of shellfish: evaluation of methods and analysis of bacteriophages and human viruses. Journal of virological methods. 2000, 89(1-2): 109-118.
Crossref - Hatib A, Hassou N, Benchekroun MN et al. The Waterborne and Food borne Viral Diseases Related to Reemerging of Poliovirus. Emerging and Reemerging Viral Pathogens. Volume 1: Fundamental and Basic Virology Aspects of Human, Animal and Plant Pathogens. 2020;44:999-1015.
Crossref - Hickey JM, Shields T, Kennedy J, Ford K. Shellfish Planting Guidelines. Division of Marine Fisheries. 2015. https://www.mass.gov/files/2017-07/shellfish-planting-guidelines_1.pdf
- Mok JS, Lee KJ, Kim PH, et al. Bacteriological quality evaluation of seawater and oysters from the Jaranman-Saryangdo area, a designated shellfish growing area in Korea: Impact of inland pollution sources. Mar Pollut Bull. 2016;108(1-2):147-154.
Crossref - Pouillot R, Van Doren JM, Woods J, et al. Meta-Analysis of the Reduction of Norovirus and Male-Specific Coliphage Concentrations in Wastewater Treatment Plants. Appl Environ Microbiol. 2015;81(14):4669-4681.
Crossref - Rosa M, Ward JE, Holohan BA, Shumway SE, Wikfors GH. Physicochemical surface properties of microalgae and their combined effects on particle selection by suspension-feeding bivalve molluscs. J Exp Mar Biol Ecol. 2017;486:59-68.
Crossref - Jofre J. Bivalve molluscs as vectors of human enteric viruses. Microbiol Aliment Nutr. 1992;10:223-228.
- Formiga-Cruz M, Allard AK, Conden-Hansson AC et al. Evaluation of Potential Indicators of Viral Contamination in Shellfish and Their Applicability to Diverse Geographical Areas. Appl Environ Microbiol. 2003;69(3):1556-1563.
Crossref - Dakki M, El Agbani MA, Qninba A. Zones humides du Maroc inscrites jusquen 2005 sur la liste de la Convention Ramsar. Trav Ins Scien. 2011:102.
- Sair AI, D’Souza DH, Moe CL, Jaykus LA. Improved detection of human enteric viruses in foods by RT-PCR. J Virol Methods. 2002b;100(1-2):57-69.
Crossref - Traore O, Arnal C, Mignotte B, Maul A, Laveran H, Billaudel S, Schwartzbrod L. Reverse transcriptase PCR detection of astrovirus, hepatitis A virus, and poliovirus in experimentally contaminated mussels: comparison of several extraction and concentration methods. Applied and Environmental Microbiology, 1998; 64(8): 3118-3122.
Crossref - LE Guyader FS, Haugarreau L, Miossec L, Dubois E, Pommepuy M. Three-Year Study To Assess Human Enteric Viruses in Shellfish. Appl Environ Microbiol. 2000;66(8):3241-3248.
Crossref - Shieh YC, Baric RS, Woods JW, Calci KR. Molecular Surveillance of Enterovirus and Norwalk-Like Virus in Oysters Relocated to a Municipal-Sewage-Impacted Gulf Estuary. Appl Environ Microbiol. 2003;69(12):7130-7136.
Crossref - Le Guyader FS, Dubois E, Menard D, Pommepuy M. Detection of Hepatitis A Virus, Rotavirus, and Enterovirus in Naturally Contaminated Shellfish and Sediment by Reverse Transcription-Seminested PCR. Appl Environ Microbiol.1994;60(10):3665-3671.
Crossref - Richards GP. Shellfish-Associated Enteric Virus Illness: Virus Localization, Disease Outbreaks and Prevention, in: Goyal, S.M., Cannon, J.L. (Eds.), Viruses in Foods, Microbiologie Alimentaire et La Securite Alimentaire. Springer International Publishing, Cham. 2016:185-207.
Crossref - Santiago CQ, Salinas CV, Bonifacio IN, Romero BLB, Ramirez EIQ. Rotavirus G2P[P] detection in fresh vegetables and oyster in Mexico City. J Food Prot. 2014;77(11):1953-1959.
Crossref - Kou X, Wu Q, Wang D, Zhang J. Simultaneous detection of norovirus and rotavirus in oysters by multiplex RT-PCR. Food Control. 2008;19(7):722-726.
Crossref - Ward RL, Bernstein DI, Young EC, et al. Human Rotavirus studies in volunteers: determination of infectious dose and serological response to infection. J Infect Dis. 1986;154(5):871-880.
Crossref - Graham DY, Dufour GR, Estes MK. Minimal infective dose of Rotavirus. Arch Virol. 1987;92(3-4):261-271.
Crossref - Dubois E. Contamination de l’environnement littoral par les rotavirus du groupe A. Detection par amplification enzymatique en chaine et analyse du polymorphisme de restriction de sequences virales dans les eaux de surfaces et les coquillages (Doctoral dissertation, University de Rennes). 1992. https://archimer.ifremer.fr/doc/00442/55376/
- Ansari SA, Springthorpe VS, Sattar SA. Survival and vehlcular spread of human rotaviruses: possible relation to seasonality of outbreaks. Rev Infect Dis. 1991;13(3):448-461.
Crossref - Gaggero A, Avendano LF, Fernandez J, Spencer E. Nosocomial transmission of rotavirus from patients admitted with diarrhea. J Clin Microbiol. 1992;30(12):3294-3297.
Crossref - Costafreda MI, Bosch A, Pinto RM. Development, Evaluation, and Standardization of a Real-Time TaqMan Reverse Transcription-PCR Assay for Quantification of Hepatitis A Virus in Clinical and Shellfish Samples. Appl Environ Microbiol. 2006;72(6):3846-3855.
Crossref - Le Guyader FS, Bon F, DeMedici D, et al. Detection of Multiple Noroviruses Associated with an International Gastroenteritis Outbreak Linked to Oyster Consumption. J Clin Microbiol. 2006;44(11):3878-3882.
Crossref - Nishida T, Nishio O, Kato M, et al. Genotyping and quantitation of noroviruses in oysters from two distinct sea areas in Japan. Microbiol Immunol. 2007;51(2):177-184.
Crossref - Le Guyader FS, Parnaudeau S, Schaeffer J, et al. Detection and Quantification of Noroviruses in Shellfish. Appl Environ Microbiol. 2009;75(3):618-624.
Crossref - El-Moqri N, El-Mellouli F, Hassou N, Benhafid M, Abouchoaib N, Etahiri S. Norovirus Detection at Oualidia Lagoon, a Moroccan Shellfish Harvesting Area, by Reverse Transcription PCR Analysis. Food Environ Virol. 2019;11(3):268-273.
Crossref - Benhafid M, Elomari N, Elqazoui N, et al. Diversity of rotavirus strains circulating in children under 5 years of age admitted to hospital for acute gastroenteritis in morocco, june 2006 to may 2009. J Med Virol. 2013;85(2):354-362.
Crossref - Matthijnssens J, Otto PH, Ciarlet M, Desselberger U, Ranst MV, Johne R. VP6 sequence-based cut-off values as a criterion for rotavirus species demarcation. Arch Virol. 2012;157(6):1177-1182.
Crossref - Krog JS, Larsen LE, Schultz AC. Enteric porcine viruses in farmed shellfish in Denmark. Int J Food Microbiol. 2014;186:105-109.
Crossref
© The Author(s) 2021. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.