ISSN: 0973-7510
E-ISSN: 2581-690X
Antibiotic resistance of pathogenic bacteria is becoming one of the most serious problems encountered by the medical community as a result of antibiotic random use. New traits of drug resistance are continuously emerging in pathogenic bacteria due to the selective pressure which is derived by antibiotic wrong use. Such resistance traits are controlled by genetic development in pathogenic bacteria. Plasmids and transposons are the most relevant DNA elements by which the traits of antibiotic resistance are both expressed in a given bacterial pathogen and transferred to other bacteria. Urinary tract infection is the most common infectious disease caused by different bacterial pathogens including Proteus spp. Which is becoming antibiotic resistant and hard to be treated, therefore, the current study was designed to survey the ability of this pathogen to resist b-lactams antibiotics especially carbapenems. The gene of oxacillinase 48 (OXA-48 bla), which is responsible for carbapenems resistance, has been detected in the plasmid DNA of 6 isolates out of 36 of multi-drug resistance Proteus spp collected over 8 months from July 2018 to March 2019. The antibiotic resistance toward 9 b-lactam antibiotics was also tested. The results showed an elevated level of resistance.
OXA-48, b-lactamases, Proteus, Antibiotic, Oxacillinases, Carbapenems, Plasmid, Resistance.
Proteus spp. is gram negative rod-shaped bacilli and one of the enterobacterial species that occur naturally in the human intestinal tract, soil, and contaminated water1. It is considered one of the opportunistic pathogens that causes different infections to the human such as urinary tract, respiratory tract, wound, otitis media, and blood infections1,2. The most common infection that caused by this bacteria is UTI as it could easily invade the urinary tract and reach kidneys especially in immuno-compromised patients. The ability of proteus in invading urinary tract is due to its strong motility ability which could be identified in laboratory by its characteristic swarming growth on petri dishes containing solid media3-5.
Bacterial infection acquired from the hospitals instruments, such as catheters and respiratory ventilation tools, is becoming a serious international problem in both developing and developed countries especially when the bacterial pathogens are progressively developing antibiotic resistance and becoming challenging toward the treatment as a result of being multi-drug resistant2,6,7.
The b-lactam antibiotics are targeted by b-lactamase enzymes produced by bacteria. These enzymes are responsible for the hydrolysis of b-lactam ring in the target antibiotic causing the later to lose its activity in inhibiting the growth of b-lactamase-producing bacteria8. b-lactamases are not showing homology at the molecular level of their structure. In other words, they are not coming from one molecular origin, therefore, they are grouped into four Amber classes: A, B, C, and D that show differences in the molecular structure, for example, classes B require zinc atom in the active site to catalyze the reaction, while active sites in the remaining classes (A, C, and D) have serine which acts as a key amino acid residue in the catalytic reactions of these classes. b-lactamases are either expressed from the plasmid or chromosomal DNA in the resistant bacteria, however, the plasmid DNA is more challenging as it provides the transferring and spreading ability of resistance genes among the pathogenic bacteria7,9.
b-lactam resistance is mainly derived by two different mechanisms: efflux pump and enzymatic mechanisms. The later is derived by b-lactamases which gave resistance toward pencillins and the early generations of cephalosporins in the past7,10,11, but the problem has extended over the past few years so that pathogenic bacteria, especially species of Enterobacteriaceae, expressed resistance against antibiotics of 4th generation cephalosporins and carbapenems that were 100% effective in bacterial infection treatment12,13.
Producing carbapenemase such as OXA betalactamases by bacteria is considered as a major problem as carbapenems antibiotics are the final option used against multi-drug resistant pathogens during the past 12 years. OXA-48-bla was firstly detected in Turkey 16 years ago expressed by imipenem resistant pathogenic Klebsiella pneumonia14. In the southern part of Asia, it was reported that Amber class B-related metallo-b-lactamase (IMP-1 and IMP-8) are expressed by Klebsiella pneumonia that was reported as carbapenems-resistant pathogen15,16.
Three different carbapenemases; KPC, NDM, and OXA-48, which belong to the amber classes A, B, and D of b-lactamases respectively, have been detected in many species of Enterobacteriaceae17,18. In the United States, Klebsiella pneumonia has been recorded to have the ability of expressing KPC-1 b-lactamase, which belongs to the class A of Abmer and gives the pathogen resistance against carbapenems19.
The present study aims to report changes in antibiotic resistance in Proteus spp which is relevant to the medical community as it highlights the development of the multi-drug resistance traits in bacteria. Molecular detection of antibiotic resistance genes, especially when genes are carried on the plasmid, provides relevant anticipation about the spreading and emergence of antibiotic resistance in pathogenic bacteria species.
Samples collection
Urine samples were collected from different clinical laboratories using sterile disposable collection cups and brought to the laboratory after few hours for culture.
Bacterial culture and identification
Centrifugation at 5000 rpm for 10 minutes were performed for urine samples using sterile centrifuge tubes. The supernatants were discarded. Pus cells presence in urine was checked by microscopic examination as a primary step to evaluate the UTI incidence. Urine samples with pus cells were cultured using brain heart infusion broth for bacterial activation. Serial dilutions were made for the brain heart broth bacterial growths and sub-cultured on blood and Macconkey agar. Colonies with Proteus spp. cultural characteristics were picked for more biochemical identification tests according to the standard manuals20.
Antibiotic susceptibility test
Discs of 9 b-lactam antibiotics including ceftriaxone, meropenem, imipenem, ceftazidime, amoxyclav, ampicillin/cloxacillin, cefixime, and cefotaxime were applied on Muller-Hinton agar cultured with Proteus spp and incubated for 20 hours at 37°C. The diameters of inhibition zones were measured after incubation.
Plasmid DNA isolation
The identified Proteus spp. isolates were sub-cultured in brain heart infusion broth overnight at 37 °C then centrifuged at 6000 rpm for 15 minutes. The alkaline lyses method for plasmid isolation was applied to the bacterial pellets using Qiagen® plasmid extraction miniprep kit. The procedure of the kit leaflet were applied.
PCR amplification and DNA electrophoresis
The isolated plasmid from each bacterial sample was used as a DNA template in PCR using two specific primers designed in this study to distinguish OXA-48 b-lactamase gene and PCR master mix from New England Biolabs®. The primers sequences are 5¹-AGCTTGATCGCCC- TCGATTT 3¹ and 5¹-GAATACCACCGTCGAGCCAG-3¹ for forward and reverse primers respectively. The PCR program was set for 31 cycles at 94°C/ 2 minutes for initial denaturation, 94°C/30 seconds for denaturation, 55°C/30 minutes for annealing, 72°C/45 minutes for elongation, and 72°C/3 minutes for final elongation. PCR products were then migrated in 1% agarose gel containing ethidium bromide at 100V for 1 hour. The DNA bands were visualized using UV transilluminator. Tris-EDTA-acetic acid buffer (TAE) of 1X concentration was used to dissolve agarose and to fill the electrophoresis tank.
One hundred and fifty urine samples were collected from clinical laboratories in Al-Najaf, Iraq. After culture, 120 urine sample gave bacterial growth, and after identification of these bacterial growths,36 Proteus spp were isolated. This indicates that proteus constitutes about 30% of the bacteria related to urinary tract infection. This is very close to the previous recent findings that recorded proteus to be a causative agent of 27% of UTIs2. It has been shown that proteus causes different infections but it is mostly (i.e. 65% of proteus) isolated from UTI13. It was revealed that the most challenged bacterial species isolated from patient suffering from UTI are E. coli and proteus21.
The results revealed that there is no antibiotic with 100% activity against proteus. This agrees with findings obtained in Sweden and Poland22. It has been reported earlier that Proteus mirablis occurs as multi-drug resistant uropathogenic bacteria expressing resistance to antibiotics of different classes including cephalosporins, aminoglycosides, and fluoroquinolones9,23.
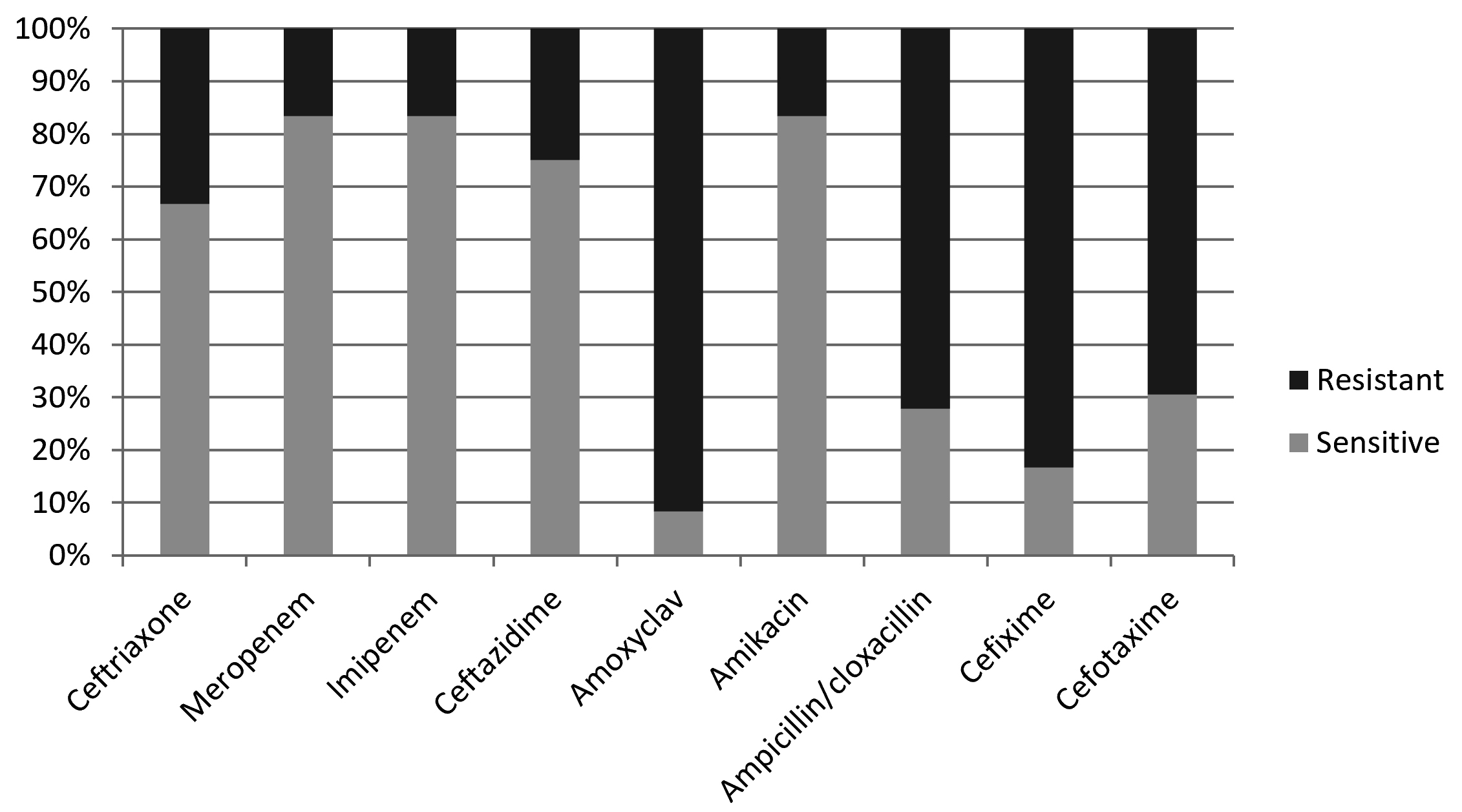
In the present study, Proteus appeared with high level of resistance toward amoxiclav, cefixime, ampicilln/cloxacillin, and cefotaxime, so that 91.7%, 83.3%, 72.2%, and 69.4% of the obtained isolates showd resistance against these b-lactams respectively (Table 1, Fig.1). On the other hand, amikacin, imipenem, and meropenem appeard to be the most active antibiotics against proteus by inhibiting the grwoth of 83.3% of the bacterial isolates, followed by ceftazidime and ceftriaxone which showed activity agaist 75% and 66.7% of the isolates respectively (Table 1, Fig. 1).
Table (1):
β-lactam antibiotic sensitivity profile of Proteus spp.
| Antibiotic | Ceftriaxone | Meropenem | Imipenem | Ceftazidime | Amoxyclav | Amikacin | Ampicillin/cloxacillin | Cefixime | Cefotaxime | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sensitivity | s | r | s | r | s | r | s | r | s | r | s | r | s | r | s | r | s | r |
| Numbers | 24 | 12 | 30 | 6 | 30 | 6 | 27 | 9 | 3 | 33 | 30 | 6 | 10 | 26 | 6 | 30 | 11 | 25 |
| Percentage | 66.7 % | 33.3 % | 83.3 % | 16.7 % | 83.3 % | 16.7 % | 75 % | 25 % | 8.3 % | 91.7 % | 83.3 % | 16.7 % | 27.8 % | 72.2 % | 16.7 % | 83.3 % | 30.6 % | 69.4 % |
The results of the present study showed that proteus is much more sensitive toward imipenem and ceftriaxone than proteus isolated in Russia that appeared more than 90% of the isolates to be completely resistant against these two drugs. Additionally, the same Russian study showed that cefotaxime, amikacin, meropenem, and ceftazidime with no activity in about 75%, 29%, 22%, and 12% of the proteus isolates respectively13. This, in general, indicates that proteus isolated in Russia is more drug resistant than its Iraqi cognate. In Swedwn, Imipenem and amikacin has been shown to be highly active agaist proteus22. It has been revealed that Proteus mirabilis occurs with notably different antibiotic resistance patterns in different countries around the world22.
Different bacterial response to the antimicrobial treatment could be due to the difference in both the type of antibiotic and how frequent they are used in different countries. The emergence of resistance to antibiotics of different classes such as the simultaneous resistance to cephalosporins and carbapenems, which is noted in this study, indicate that resistance traits are transferred continuously among the bacterial pathogens. Therefore, diagnosing the co-existance of resistance toward different antibiotics highlights the propensity of the bacteria in arising multi-drug resistance.
Using specific primers designed in this study, the PCR genetic analysis of plasmid DNA isolated from Proteus spp revealed that 6 out of 36 bacterial isolates (16.7%) hold the gene of OXA-48-b-lactamase (Fig. 2). These 6 isolates showed resistance to all antibiotics used in this study including carbapenems (meropenem and imipenem), while the other isolates that showed negative PCR amplification of OXA-48-b-lactamase gene were sensitive toward carbapenems. This indicates that the key reason behind the arbapenems resistance in the PCR positive isolates is the OXA-48 enzyme.
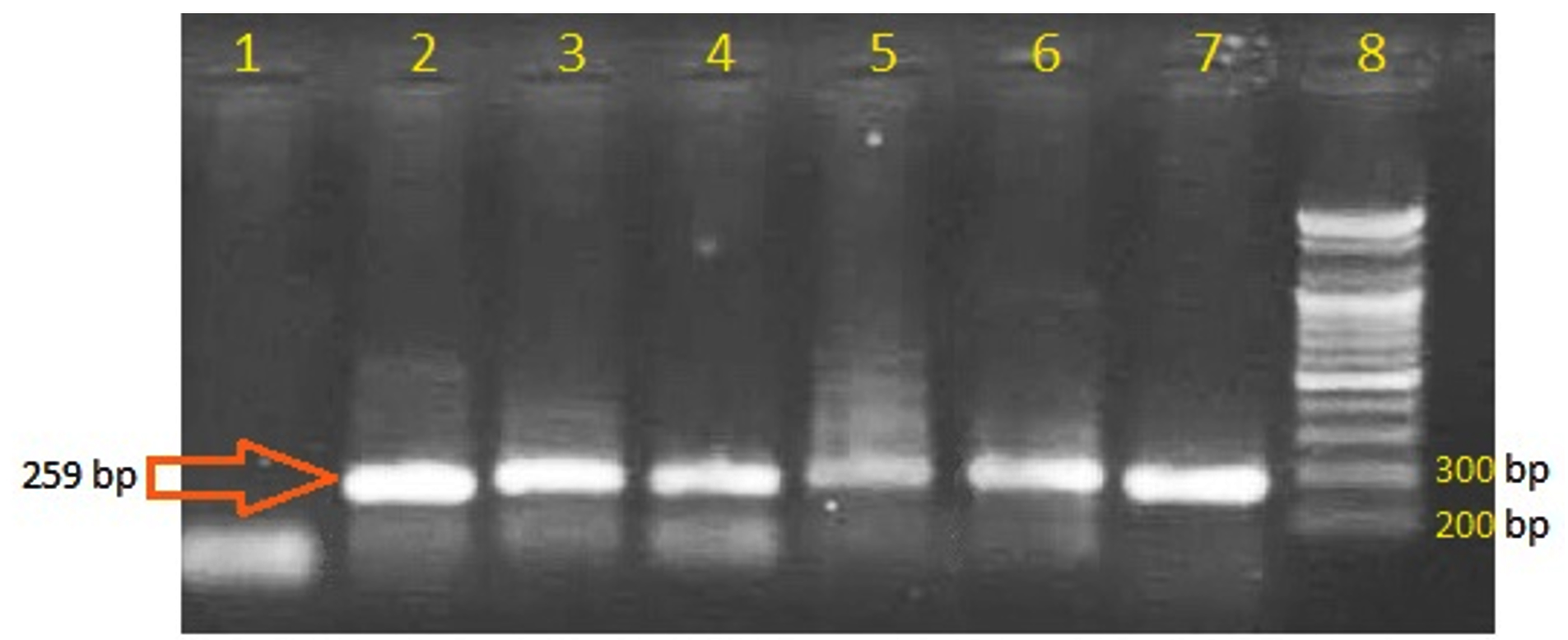 Fig. 2. PCR amplification of OXA-48-β-lactamase. Lanes 2-7 are positive amplification bands with the expected size indicating the presence of the OXA-48 gene. Lane 1 is negative control. Lane 8 is DNA ladder.
Fig. 2. PCR amplification of OXA-48-β-lactamase. Lanes 2-7 are positive amplification bands with the expected size indicating the presence of the OXA-48 gene. Lane 1 is negative control. Lane 8 is DNA ladder. The plasmid DNA was the target in the present study as it provides the mobile genetic element of transferring the traits of antibiotic resistance among the pathogenic bacteria, therefore, it is more relevant than the chromosomal DNA to be studied in reports of antibiotic resistance emergence. The positive results of holding OXA-48 gene on the plasmid of proteus, which is not recorded before in Iraq, highlights the emergence of carbapenems resistance that could be widely spread among the species of Enterobacteriaceae.
The genetic expression ability of OXA-48 is not exclusive for certain bacterial species but it is common in many Enterobacterial pathogens including proteus and many other bacteria such as klebsiella, enterobacter, morganella, citrobacter, and serratia as reported by many studies24-26.
Turkey as one of the middle east countries is considered as one of the endemic regions of OXA-48-mediated antibiotic resistance. This may explain the emergence of such resistance trait in Iraq. Northern African countries and India have also been reported to be endemic regions of OXA-48. Additionally, the incidence of OXA-48 bacterial producers was reported in China, Australia, and many European and south American countries27.
The wide outbreak of OXA-48 and other genes of similar b-lactamases could be explained as these genes were reported to be maintained on a 6200 bp IncL-related plasmid which is easily and more frequently transferred among bacterial hosts by conjugation due to the lack of conjugation controlling mechanism in such plasmids that results from the insertional transposition of oxacillinases genes in the region of the gene of repressor protein (tir protein) that is responsible for the conjugation control in such kind of plasmids28. It has been revealed that plasmid-mediated OXA-48- b-lactamase is most likely transferable horizontally and clonally among bacterial cells 24.
Oxacillinases exhibit high levels of similarity in their protein primary structure except some few amino acid residues changes which have been recorded29,30. Similarity in primary structure leads to similarity in the tertiary structure and subsequently functional properties of these enzymes. Therefore, OXA-b-lactamases (including OXA-48) are all decribed as weak enzymes that do not catalyse their own biochemical reactions without the interference of other assisting factors such as the reduction of bacterial cell barrier permeability which is emerged in bacteria after several mutational changes in the porin protein31. This indicates that carbapenems resistance of proteus isolates obtained in the current study (Table 1) has emerged as a result of multi-factorial combination of cellular adaptation mechanisms including not only OXA-48 production but also adaptive mutational changes that lead to the decreased cellular permeability.
Acknowledgements
None.
Funding
None
Data Availability
All datasets generated or analyzed during this study are included in the manuscript.
Ethics Statement
This article does not contain any studies with human participants or animals performed by any of the authors.
- Guentzel MN. Escherichia, klebsiella, enterobacter, serratia, citrobacter, and proteus. Medical Microbiology. 4th edition: University of Texas Medical Branch at Galveston; 1996.
- Al-Shamarti MJ, Hussein AA, AL-Luhaiby AI. The Relationship Between the Type of Infection and Antibiotic Resistance. J. Pure Appl. Microbiol., 2018; 12(2): 845-854.
Crossref - Rauprich O, Matsushita M, Weijer CJ, Siegert F, Esipov SE, Shapiro JA. Periodic phenomena in Proteus mirabilis swarm colony development. J. Bacteriol., 1996; 178(22): 6525-6538.
Crossref - Matsuyama T, Takagi Y, Nakagawa Y, Itoh H, Wakita J, Matsushita M. Dynamic Aspects of the Structured Cell Population in a Swarming Colony of Proteus mirabilis. J. Bacteriol., 2000; 182(2): 385-393.
Crossref - Ryan KJ, Ray CG. Medical microbiology. McGraw Hill. 2004;4:370.
- Rosenthal VD, Bijie H, Maki DG, et al. International Nosocomial Infection Control Consortium (INICC) report, data summary of 36 countries, for 2004-2009. American Journal of Infection Control, 2012; 40(5): 396-407.
Crossref - Garcia-Rodriguez J, Jones R, Group, Study TMP. Antimicrobial resistance in gram-negative isolates from European intensive care units: data from the Meropenem Yearly Susceptibility Test Information Collection (MYSTIC) programme. Journal of chemotherapy, 2002; 14(1):25-32.
Crossref - Empel J, Baraniak A, Literacka E, et al. Molecular survey of b-lactamases conferring resistance to newer b-lactams in Enterobacteriaceae isolates from Polish hospitals. Antimicrobial Agents and Chemotherapy, 2008; 52(7): 2449-2454.
Crossref - Pagani L, Migliavacca R, Pallecchi L, et al. Emerging extended-spectrum b-lactamases in Proteus mirabilis. J. Cli. Microbiol., 2002; 40(4): 1549-1552.
Crossref - Al-Shamarti M. molecular evaluation of B-lactam resistance genes in klebsiella spp isolated from clinical cases in Al-Najaf province. Department of Biology. University of Kufa, 2010.
- ALshamarti MJ, AL-Muhnna A. Molecular detection of AmpC Gene encoding antibiotic resistance among Klebsiella SPP. isolated from different infections. Al-Kufa Journal for Biology, 2011; 3(1): 1-9.
- Birgand G, Armand-Lefevre L, Lepainteur M, et al. Introduction of highly resistant bacteria into a hospital via patients repatriated or recently hospitalized in a foreign country. Clinical Microb. and Infection, 2014; 20(11): O887-O890.
Crossref - Fursova NK, Astashkin EI, Knyazeva AI, et al. The spread of bla OXA-48 and bla OXA-244 carbapenemase genes among Klebsiella pneumoniae, Proteus mirabilis and Enterobacter spp. isolated in Moscow, Russia. Annals of clinical Microbiology and Antimicrobials. 2015; 14(1): 46.
Crossref - Poirel L, Hיritier C, Tol n V, Nordmann P. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrobial Agents and Chemotherapy. 2004; 48(1): 15-22.
Crossref - Koh T, Babini G, Woodford N, Sng L, Hall L, Livermore D. Carbapenem-hydrolysing IMP-1 b-lactamase in Klebsiella pneumoniae from Singapore. The Lancet, 1999; 353(9170): 2162.
Crossref - Yan J-J, Ko W-C, Wu J-J. Identification of a plasmid encoding SHV-12, TEM-1, and a variant of IMP-2 metallo-b-lactamase, IMP-8, from a clinical isolate of Klebsiella pneumoniae. Antimicrobial Agents and Chemotherapy. 2001; 45(8): 2368-2371.
Crossref - Ambler R. The nature of 1-lactamases. Philos. Trans. R. Soc. London B. Biol. Sci., 1980; 289: 321-331.
Crossref - Ageevets VA, Partina IV, Lisitsyna ES, et al. Emergence of carbapenemase-producing Gram-negative bacteria in Saint Petersburg, Russia. International Journal of Antimicrobial Agents. 2014; 44(2): 152-155.
Crossref - Yigit H, Queenan AM, Anderson GJ, et al. Novel carbapenem-hydrolyzing b-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrobial Agents and Chemotherapy. 2001; 45(4): 1151-1161.
Crossref - Mac Faddin J. Biochemical Tests for Identification of Medical. Bacteria. Williams and Wilkins. London. 2000.
- Lipsitch M, Bergstrom CT, Levin BR. The epidemiology of antibiotic resistance in hospitals: paradoxes and prescriptions. Proceedings of the National Academy of Sciences, 2000; 97(4): 1938-1943.
Crossref - Adamus-Bialek W, Zajac E, Parniewski P, Kaca W. Comparison of antibiotic resistance patterns in collections of Escherichia coli and Proteus mirabilis uropathogenic strains. Molecular Biology Reports, 2013; 40(4): 3429-3435.
Crossref - Khan AU, Musharraf A. Plasmid-mediated multiple antibiotic resistance in Proteus mirabilis isolated from patients with urinary tract infection. Medical science monitor: Iinternational Medical Journal of Experimental and Clinical Research, 2004; 10(11): CR598-602.
- Arana D, Saez D, Garcםa-Hierro P, et al. Concurrent interspecies and clonal dissemination of OXA-48 carbapenemase. Clinical Microbiology and Infection. 2015; 21(2): 148. e141-148. e144.
Crossref - Hammoudi D, Moubareck CA, Aires J, et al. Countrywide spread of OXA-48 carbapenemase in Lebanon: surveillance and genetic characterization of carbapenem-non-susceptible Enterobacteriaceae in 10 hospitals over a one-year period. International Journal of Infectious Diseases. 2014; 29: 139-144.
Crossref - Chen L, Al Laham N, Chavda KD, et al. First report of an OXA-48-producing multidrug-resistant Proteus mirabilis strain from Gaza, Palestine. Antimicrobial Agents and Chemotherapy, 2015; 59(7): 4305-4307.
Crossref - Nordmann P, Poirel L. The difficult-to-control spread of carbapenemase producers among Enterobacteriaceae worldwide. Clinical Microbiology and Infection, 2014; 20(9): 821-830.
Crossref - Potron A, Poirel L, Nordmann P. Derepressed transfer properties leading to the efficient spread of the plasmid encoding carbapenemase OXA-48. Antimicrobial Agents and Chemotherapy, 2014; 58(1):467-471.
Crossref - Martםnez-Martםnez L, Gonzבlez-Lףpez JJ. Carbapenemases in Enterobacteriaceae: types and molecular epidemiology. Enfermedades Infecciosas y Microbiologia Clinica. 2014; 32: 4-9.
Crossref - Oteo J, Hernבndez JM, Espasa M, et al. Emergence of OXA-48-producing Klebsiella pneumoniae and the novel carbapenemases OXA-244 and OXA-245 in Spain. Journal of Antimicrobial Chemotherapy, 2012; 68(2): 317-321.
Crossref - Poirel L, Naas T, Nordmann P. Diversity, epidemiology, and genetics of class D b-lactamases. Antimicrobial Agents and Chemotherapy, 2010; 54(1): 24-38.
Crossref
© The Author(s) 2019. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.