ISSN: 0973-7510
E-ISSN: 2581-690X
Antibiotic resistance has become a serious global threat, mainly due to misuse, overuse of antibiotics and non-compliance with infection control protocol. Superbugs are multidrug-resistant (MDR) and extended drug-resistant (XDR) bacteria, mainly Klebsiella pneumoniae and Escherichia coli from the Enterobacteriaceae family, which cause opportunistic infections and raise death rates and hospital expenditures. The present study was conducted at a tertiary care teaching hospital to study the epidemiology and molecular detection of carbapenem-resistant K. pneumoniae isolated from various clinical specimens. 240 K. pneumoniae isolates were collected from January 2020 to December 2021 at the Bacteriology laboratory, Index Medical College and Hospital, Indore. All isolates were analyzed for carbapenem resistance by the conventional disc diffusion method. All carbapenem-resistant isolates were tested for carbapenemase production using the phenotypic double-disk synergy test (DDST) and modified Hodge test (MHT) as per 2020 CLSI guidelines. All isolates were negative by phenotypic methods, further confirmed by conventional PCR to detect the gene responsible for carbapenemase production. 240 isolates of K. pneumoniae were included during the study periods. Out of 240 isolates, 102 isolates were found resistant to carbapenem drugs. All 102 isolates were confirmed carbapenemase and MBL producers by MHT and DDST tests. Among 102, 60 isolates were found to be MBL producers negative by MHT and DDST tests. Sixty phenotypic negative carbapenem-resistant isolates were tested by conventional PCR. One or more carbapenemase genes were detected in 61.0% of isolates. The blaKPC was detected in 13/60 (21%) isolates, followed by blaNDM 10/60 (16%) isolates, followed by blaVIM in 6/60(10%), blaOXA-48 in 5/60 (8%) and blaIMP in 3/60(5%) isolates. K. pneumoniae produces carbapenemase, which enhances resistance to the carbapenem class of antibiotics. The simultaneous detection of these resistance genes expressed by Klebsiella pneumoniae might be managed by early detection and adhering to antibiotic policies that limit the use of antibiotics.
Double Disk Synergy Test (DDST), Modified Hodge test (MHT), Metallo-beta-Lactames (MBL), Polymerase Chain Reaction
Antibiotic resistance has become a serious global threat, this is mainly due to misuse, overuse of antibiotics and non-compliance with infection control protocols.1-3 The multidrug resistant (MDR) and extensive drug resistant (XDR), bacteria are called as ‘Superbug’ especially seen in Enterobacteriaceae family with Klebsiella pneumoniae followed by Escherichia coli which causes opportunistic infections and leads to increase in death rate which causes increase in expenses of hospital related costs.4-6 In 2017, World Health Organization has classify carbapenem-resistant Enterobacteriaceae (CRE), on the global priority among top 10 global public health threats facing humanity.1,4,7,8
CRE is due to Beta-lactamases, Carbapenemeses, mutation in bacteria and efflux pumps, it alters the expression and functions of porins and proteins that binds with penicillin and also combinations of these mechanisms lead to high levels of carbapenem resistance is seen in K. pneumoniae.9,10 CRE classified into three different molecular classes like A, B and D, its examples are Serines carbapenemes, such as K. pneumoniae Carbapenemes, (KPC) are examples of carbapenemases in class A. Metallobactam lactamases (MBLs), like Verona integron encoded MBL, (VLM), New Delhi MBL (NDM), and imipenemase (IMP) are examples of class B, and OXA carbapenemases like blaOXA-48 are examples of molecular class D.9-11 High prevalence of CRE is reported from southern Europe and Asia than in other parts of the world.12 CRE is mostly encounter in nosocomial isolates than community isolates.
Center for Disease Dynamics, Economics and policy 2021, stated in Indian scenario there is the markedly increase in K pneumoniae, it was 24% in 2008 and it is 59% in 2017. It is observed due to inadequate medical intervention, various co-morbidities and over use of antibiotics.13
For the detection of KPCs, several phenotypic tests have been established.14,15 Presently Modified Hodge test (MHT) is preferred and accepted as accurate and sensitive method for detection of carbapenemase which is approved by CLSI guidelines.16,17 MHT cannot be used as a confirmatory test for detection of the KPCs because of difficult clarification and false positive results.18,19 KPCs isolates producing AmpC and CTX-M β-lactamase are showing commonly as false positive results.20
For the identification of resistance conferring genes, gold standard method preferred is Polymerase Chain Reaction (PCR), which amplifies a specific nucleic acid target, to obtain a million or more copies which can then be easily detected by using nucleic acid staining techniques.21
In spite of high disease burden, limited Indian studies describe mechanisms of resistance caused due to K. Pneumoniae isolates in MDR, which highlights the requirement of comprehensive epidemiological surveillance results.22 Therefore, the present study was conducted at a tertiary care teaching hospital with an aim to study epidemiology, antimicrobial susceptibility profile of carbapenem-resistant K. pneumoniae isolated from various clinical specimens.
A total of 240 Klebsiella pneumoniae isolates was collected from Index Medical College and Hospital, Indore, during the period January 2020 to December 2021. From the clinical specimens such as urine, CSF, wounds, blood, sputum; bacteria was identified using by standard Conventional methods.
Culture Media and Chemicals
All required culture medium, antibiotics and chemicals (analytical grade) were procured from HIMEDIA Pvt. Ltd. Mumbai, India, and molecular testing reagents using QIAamp DNA mini kit (Qiagen India Pvt. Ltd.) for bacterial genomic DNA extraction and master mix (HotStarTaq Master Mix Kit(Qiagen Cat. No. 203443) were used in this study.
Antibiotic susceptibility test
By using Kirby-Bauer method, antibiotic susceptibility test was done for all isolates according to CLSI standards guidelines 2020.23 Selection of antibiotics was done as per CLSI 2020 guidelines. Following antibiotic discs used in the study were Amikacin (30µg), Gentamicin (10µg), Ceftazidime (30µg), Ciprofloxacin (5µg), Imipenem (10µg), Meropenem (10µg), Aztreonam (30µg), Piperacillin / Tazobactum (100/10µg). Amoxiclave (30µg), Cefixime(5µg), Ceftriaxone (30µg), Cefuroxime (30µg), Cefepime (30µg), Tetracycline (30µg), Trimethoprim (5µg) Cefotaxime (30µg), Ampicillin (10µg), Nitrofuration (300µg).
Carbapenemase screening
Phenotypic Methods
By performing Modified Hodge test (MHT), all isolates were screened for determination of carbapenemase production. To detect MBL (Metallo-beta-Lactames) production, a double disk synergy test (DDST) was done as per the CLSI guidelines 2020.23
Genotypic Methods
The genes responsible for carbapenemase production may be initiating the resistance to carbapenem groups antibiotics were detected by genotypic methods by using QIAamp DNA mini kit (Qiagen India Pvt. Ltd.) for bacterial genomic DNA extraction and master mix (HotStarTaq Master Mix Kit(Qiagen Cat. No. 203443) With the help of above this kit performed conventional PCR to detect the presence of genes such as NDM-1, OXA-48, KPC, VIM and IMP in K. pneumoniae isolates. The primers used in this study are enlisted below (Table 1).
Table (1):
Primers are used for the detection of target genes.
Gene target |
Primers sequence (5’-3’) |
Product size (bp) |
|---|---|---|
NDM – F NDM –R |
GGGCAGTCGCTTCCAACGGT GTAGTGCTCAGTGTCGGCAT |
188 |
OXA -48F OXA -48 R |
TTGGTGGCATCGATTATCGG GAGCACTTCTTTTGTGATGGC |
390 |
VIM-F VIM-R |
GATGGTGTTTGGTCGCATA CGAATGCGCAGCACCAG |
743 |
IMP-F IMP-R |
GGAATAGAGTGGCTTAATTCTC CCAAACCACTACGTTATCT |
475 |
KPC F KPC R |
TGTCACTGTATCGCCGTC CTCAGTGCTCTACAGAAAACC |
1000 |
Master mix and PCR Cycling conditions were prepared as per kit standard protocol. Electrophoresis were used for the detection of amplicon band, using 2% agarose gel performed at 60-90 V for 30-45 mins. The gel documentation was done by using gel doc system (Bio Era, India). 100 bp DNA ladder (QiagenGelPilot) was used parallel to test for marking of molecular weight.
A Total of 240 isolates of K. pneumoniae isolates (126 in patients and 114 outpatients) were used in the study, among 108 isolates were collected from male and 132 from female participants. Antibiotic resistant were showed in Table 2.
Table (2):
Antibiotic-resistant pattern for K. pneumoniae isolates.
Antibiotic |
Total N= 240 (%) |
|---|---|
Imipenem |
102 (41.7) |
Meropenem |
102(41.7) |
Ampicillin |
240 (100) |
Amoxyclav |
217 (90.4) |
Ceftazidime |
240 (100) |
Ceftriaxone |
226 (94.1) |
Cefotaxime |
233 (97.1) |
Cefuroxime |
213(88.7) |
Aztreonam |
213 (88.7) |
Cefixime |
198(82.5) |
Piperacillin+Tazobactum |
220 (91.7) |
Tetracycline |
198 (82.5) |
Cefepime |
224 (93.3) |
Trimethoprim |
211(87.9) |
Ciprofloxacin |
194 (80.8) |
Gentamicin |
153 (63.8) |
Amikacin |
152 (63.3) |
Nitrofurantoin |
38(15.8) |
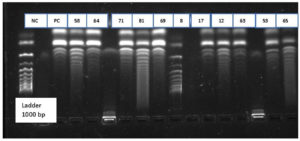
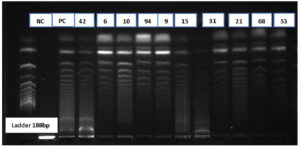
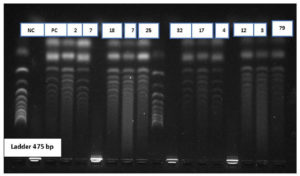
A total of 102 isolates observe to be resistant to carbapenem drugs. These were confirmed by phenotypic methods MHT and DDST for carbapenemase producers. Among 102 isolates, 60 isolates were negative to phenotypic methods. These 60 isolates were further confirmed by the molecular method using conventional PCR to detection of resistant genes. KPC Genes in resistant isolate shows in Figure 1 as the negative control (NC) Positive control (PC) and showing samples numbers with DNA Ladder 1000bp. NDM Genes in resistant isolate shows in Figure 2 as the negative control (NC) Positive control (PC) and showing samples numbers with DNA Ladder 188bp. IMP Genes in resistant isolate shows in Figure 3 as the negative control (NC) Positive control (PC) and showing samples numbers with DNA Ladder 475bp. Of 60 Carbapenem-resistant isolates, 37(61%) were positive for one and multiple targeted genes. blaKPC was detected in 21% of isolates, followed by blaNDM 16.0%, blaVIM in 10.0%, blaOXA-48 8.0% and blaIMP in 5% (Table 3).
Table (3):
Frequency distribution of Carbapenemase genes family in various clinical specimens.
| Specimen type | Frequency of carbapenemase genes | No. | ||||
|---|---|---|---|---|---|---|
| blaNDM | blaKPC | blaOXA-48 | blaVIM | blaIMP | ||
| Blood | 3 | 4 | – | 2 | 1 | 10 |
| Urine | 1 | 3 | 2 | 1 | 1 | 8 |
| Wound | 3 | 3 | – | – | 1 | 7 |
| Sputum | 1 | 2 | 1 | 2 | – | 6 |
| CSF | 2 | 1 | 2 | 1 | – | 6 |
| Total | 10 (16%) | 13 (21%) | 5 (8%) | 6 (10%) | 3 (5%) | 37 (61%) |
In recent years, molecular diagnostic techniques have become a game changer for clinical laboratories of all sizes. Molecular diagnostics offer more powerful tools for earlier and more accurate detection of various diseases, including infectious diseases.12
Today, molecular diagnostic methods testing has become more routine, mainly due to the development of automated instrument systems that provide accurate and precise results utilizing polymerase chain reaction (PCR) and other molecular-based technologies for identification and DNA/RNA measurement of infectious pathogens, tumours, and human genes.24
Various genetic mechanisms, including efflux pumps, altered function and expression of porins and penicillin-binding proteins (PBPs), are considered to be involved in carbapenem resistance in Enterobacteriaceae. However, several mechanisms are yet to be identified or discovered.25
A golden rule for building any clinical laboratory capacity in resource-limited settings is selecting the most appropriate workable, affordable, and sustained techniques. In the present study, an in-depth molecular characterization of K. pneumoniae was conducted by in-house conventional PCR. Nucleic acid-based assays for the detection of antimicrobial resistance may offer advantages over phenotypic methods.26,27
The inclusion criterion for molecular detection of carbapenem resistant genes was K. pneumoniae demonstrating carbapenem resistance by the E-test method.27 For conventional PCR, the blaKPC, blaNDM, blaVIM, blaIMP and blaOXA were done using primers. Out of 60 K. pneumoniae isolates, from sequencing analysis, only three genes, namely blaKPC, blaIMP, and blaNDM were detected.
In this study, the high prevalence of blaKPC gene13 (21%) was isolated, followed by blaNDM was detected in 10(16%), blaVIM in 6 (10%), blaOXA-48 in 5 (8%) and blaIMP in 3 (5%). Various researchers increasingly reported similar genes responsible for carbapenem resistance at a rapid velocity in K. pneumoniae. Baran et al.,28 reported detection of carbapenemase-encoding genes in 81.7% (94/115) isolates. The genes detected in their study were blaKPC (78.3%), blaNDM-1 (0.9%), and blaSME (2.6%). This study reported the increased isolation of carbapenem resistant K. pneumoniae and KPC as the commonest gene responsible for resistance.
In the study by Hamzan et al.,29 only two genes, bla IMP4 and blaNDM1 were detected in K. pneumoniae using conventional PCR. Mohanty et al. 30 reported the highest frequency of NDM 1 in 65.6% and OXA 48 in 24.7%, OXA 181 23.6%, VIM in 6.4% and KPC in 2.1% of K. pneumoniae isolates. In their study blaIMP gene was not found in any of the isolates by using conventional PCR. A major advantage of conventional PCR is its cost effectiveness and ready access to conventional thermo cyclers that almost all research facilities. Veeraraghavan et al. 31 to performed multiplex PCR for detection of resistance genes encoding α-lactam resistance. In their study, among the carbapenemases co-expression of blaNDM and blaOXA48 -like was observed in 28%, blaNDM in 19%, blaOXA48 -like in 13% and blaKPC was not found. In the study of Vivan et al.,32 the isolates that were phenotypically screened for carbapenemase production were subjected for genotypic confirmation by PCR (PCR) for KPC, metallo-β-lactamases, OXA-48, and extended-spectrum beta-lactamase genes. PCR analysis demonstrated that all isolates carried blaKPC genes and sequencing showed that all strains belonged to KPC-2 subtype.
In the study by Pourgholi et al.,33 blaKPC2 has been detected in 8 out of 17 carbapenem resistant K. pneumoniae isolates from a tertiary teaching hospital. Ahmad et al.34 from Aligarh, performed molecular characterization of novel sequence type of carbapenem-resistant New Delhi metallo-β-lactamase-1-producing K. pneumoniae isolated from NICU. In their study, all 17 isolates were found to carry blaNDM (13 blaNDM-1, 1 blaNDM-4 and 3 blaNDM-5), seven isolates carried blaOXA-48, 13 isolates had blaCTX-M-15, seven isolates carried blaCMY-1 and five isolates were found to carry blaSHV-1. Galani et al.35 detected 300 carbapenem resistant K. pneumoniae strains in hospitals across Greece and found KPC-2 (66.7%), NDM (16.7 %), VIM (7%) and OXA-48 (4%) whereas 14 strains carried both KPC and VIM (4.7%), two strains carried both NDM and OXA (0.7%) and one strain carried both KPC and OXA (03%) resistance genes. Shankar et al.,36 observed that 60 % of the isolates co-produced blaSHV, blaTEM and blaCTX- M-15. blaOXA48 -like was the predominant carbapenemase gene produced in 71 % followed by co-production of blaNDM and blaOXA48 in 11 per cent, blaNDM in 7% and blaKPC 3% and four isolates did not produce any of the carbapenemases genes tested.
Bhaskar et al.37 studied 165 carbapenem resistant K. pneumoniae isolates. In their study, 9.7% were positive for blaNDM-1 and these isolates were also found to be positive for one or more bla genes. Co-carriage of AmpC in ESBL and carbapenem resistant isolates were 7.8% and 3.6%, respectively and were negative for blaKPC genes. Han et al.38 reported the KPC-2 gene as the most commonly detected followed by the CTX-M9 and OmpK.36 However, in their study, IPM-4, NDM-1 and OXA-48 were not detected.
The epidemiology of carbapenem resistant K. pneumoniae was studied by Lau et al.39 using molecular method. This study included 63 carbapenem resistant strains of K. pnuemoniae. Carbapenemase genes were detected in 55 isolates, with blaOXA-48 (63.5%) as the predominant carbapenemase gene, followed by blaNDM (36.5%). Alizadeh et al.40 reported that blaVIM-1 as the most prevalent gene followed by blaIMP-1 and blaNDM-1. This study suggested that PCR could be considered a suitable tool for the rapid identification of strains with high epidemic potential that may be required for local outbreak studies. Li et al.41 studied antimicrobial susceptibility profile, molecular characteristics, plasmid and integron-associated analysis, genetic environments of blaKPC-2 and blaNDM-1 of 66 strains of carbapenem-resistant K. pneumoniae isolated from BSIs. In their study, 3 strains were identified co-carrying blaNDM and blaIMP genes, including two isolates with blaNDM-1 and blaIMP-4 and one with blaNDM-5 and blaIMP-4.
Kazi et al.42 from Mumbai, reported that predominance of blaNDM1 gene in their study by using multiplex PCR. These authors concluded that PCR is important for rapid detection of genes responsible for drug resistance and also highlighted the importance of implementation of strict infection control measures and contact precautions to prevent spreading of blaNDM1 mediated resistance in health-care setup. Bhatia et al.43 studied 29 MDR K. pneumoniae isolates obtained from various clinical samples. These authors reported carbapenem resistance in 27 isolates. Carbapenem resistance was mainly encoded by OXA-48-like genes (21/27 [77.8%]) and all isolates had a varied arsenal of resistance genes to different antibiotic classes. Bhatia et al. highlighted the continuous need for genomic surveillance of MDR bacteria for developing treatment guidelines based on integrating phenotypic and molecular methods. In the study of Bhatt et al.44 Out of 150 phenotypically confirmed carbapenemase-producing isolates, blaNDM gene was found in 85, blaVIM in 32, and blaIMP in 22 isolates. blaOXA-48 and blaKPC genes were not found in any isolate. Moreover, there were 19 isolates, in which no gene was detected.
In this study, carbapenem resistance was confirmed in 60 (25%) out of 240 K. Pneumoniae isolated from different clinical specimens was done with polymerase chain reaction blaKPC, blaIMP, and blaNDM was the genes found to be associated with carbapenem resistance K. pneumoniae. As carbapenem resistance was high, this study highlights the importance of strict compliance with infection prevention and control measures, rational use of antimicrobials, and active surveillance for the presence of carbapenemase.
K. pneumoniae increases resistance to the carbapenem group of antimicrobials by producing carbapenemase. For the reservoir of resistance, carbapenemase non-producing isolates act to MBL and KPC production, which were associated with increased mortality and morbidity and can spread within healthcare settings and communities. Simultaneous detection of these resistance patterns of MBL and KPC-producing Klebsiella isolates can be controlled by early detection and antibiotic policies by curtailing the injudicious use of antibiotics. The cross-transmission of multidrug-resistant organisms would prevent the emergence and implementation of antimicrobial stewardship.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
ETHICS STATEMENT
This study was approved by the Institutional Ethics Committee, Malwancal University, Indore, India, with reference number MU/Research/EC/Ph.D/2018/13.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
- World Health Organization.Global Action Plan on Antibiotic Resistance.WHO Press: Geneva, Switzerland, 2015.
- Carlet J, Jarlier V, Harbarth S, Voss A, Goossens H, Pittet D. Participants of the 3rd World Healthcare-Associated Infections Forum. Ready for a world without antibiotics? The Pensieres Antibiotic Resistance Call to Action. Antimicrob Resist Infect Control. 2012;1:11.
Crossref - World Health Organization. Antimicrobial Resistance: Global Report on Surveillance; WHO Press: Geneva, Switzerland, 2014.
- Marr CM, Russo TA. Hyper virulent Klebsiella pneumoniae: a new public health threat. Expert Rev Anti Infect Ther. 2019;17(2):71-73.
Crossref - Yard CW. The drug-resistant bacteria that pose the greatest health threats. Nature. 2017;543:15.
Crossref - Hyun M, Noh CI, Ryu SY, Kim HA. Changing trends in clinical characteristics and antibiotic susceptibility of Klebsiella pneumoniae bacteremia. Korean J Intern Med. 2018;33(3):595-603.
Crossref - Kang J, Yi J, Ko M, Lee S, Lee J, Kim K. Prevalence and risk factors of Carbapenem-resistant Enterobacteriaceae Acquisition in an Emergency Intensive Care Unit in a Tertiary Hospital in Korea: a Case-Control Study. J Korean Med Sci. 2019;34(18):e140.
Crossref - Zhang Y, Wang Q, Yin Y, et al. Epidemiology of carbapenem-resistant Enterobacteriaceae infections: report from the China CRE network. Antimicrob Agents Chemother. 2018;62(2):e01882.
Crossref - Gao F, Xiong Z, Liang B, et al. Molecular Characterization and Epidemiology of Carbapenem-Resistant Enterobacteriaceae Isolated from Pediatric Patients in Guangzhou, Southern China. Can J Infect Dis Med Microbiol. 2023;4762143.
Crossref - Canton R, Akova M, Carmeli Y, et al. Rapid evolution and spread of carbapenemases among Enterobacteriaceae in Europe. Clin Microbiol Infect. 2012;18(5):413-431.
Crossref - Grover SS, Sharma M, Pasha ST, Singh G, Lal S. Antimicrobial susceptibility pattern and prevalence of extended spectrum beta-lactamase (ESBLs) producing strains of Klebsiella pneumoniae from a major hospital in New Delhi. The Journal of Communicable Diseases. 2004;36(1):17-26
- Iskandar K, Molinier L, Hallit S, et al. Surveillance of antimicrobial resistance in low-and middle-income countries: a scattered picture. Antimicrobial Resistance & Infection Control. 2021;10(1):1-9.
- The Center for Disease Dynamics, Economics & Policy. Resistance Map: India. 2021. https://resistancemap.cddep.org/CountryPage.php?countryId=17&country=India. Date accessed: 13th May, 2021
- Hirsch EB, Tam VH. Detection and treatment options for Klebsiella pneumonia carbapenemases (KPCs): an emerging cause of multidrug-resistant infection. J Antimicrob Chemother. 2010;65(6):1119-1125.
Crossref - Vatopoulos A. High rates of Metallo-b-lactamase producing Klebsiella pneumonia in Greece-a review of the current evidence. Euro Surveill. 2008;13(4):8023.
- Cury AP, Andreazzi D, Maffucci M, Caiaffa-Junior HH, Rossi F. The modified Hodge test is a useful tool for ruling out Klebsiella pneumoniae carbapenemase. Clinics. 2012;67(12):1427-1431.
Crossref - Chen S, Hu F, Xu X, et al. High Prevalence of KPC-2-Type Carbapenemase Coupled with CTX-M-Type Extended-Spectrum-Lactamases in Carbapenem-Resistant Klebsiella pneumoniae in a Teaching Hospital in China. Antimicrob Agents Chemother. 2011(5):2493-2494.
Crossref - Pasteran F, Mendez T, Guerriero L, Rapoport M, Corso A. Sensitive screening tests for suspected class a carbapenemase production in species of Enterobacteriaceae. J Clin Microbiol. 2009;47(6):1631-1639.
Crossref - Lee K, Chong Y, Shin HB, Yong D, Yum JH. Modified Hodge and EDTA-disk synergy tests to screen metallo-b-lactamase producing strains of Pseudomonas and Acinetobacter species. Clin Microbiol Infect. 2001;7:88-91.
Crossref - Carvalhaes CG, Picao RC, Nicoletti AG. Cloverleaf test (modified Hodge test) for detecting carbapenemase production in Klebsiella pneumoniae: be aware of false positive results. J Antimicrob Chemother. 2010;65(2):249-251.
Crossref - Naaber P, Rudzko R, Ivanova M, Koressaar T, Pavelkovich A, Ratnik K. Comparison of Coris rapid test with other methods for carbapenemases detection in Klebsiella pneumoniae. in Proceedings of the 35th Annual Meeting of NSCMID; 19 – 22 August 2018, (Reykjavik).
- Srivastava P, Bisht D, Kumar A, Tripathi A. Prevalence of Carbapenem-Resistant Escherichia coli and Klebsiella pneumoniae in Rural Uttar Pradesh. Journal of Datta Meghe Institute of Medical Sciences University. 2022;17(3):584
- CLSI Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; 22nd Informational Supplement. CLSI Document M100-S22. Wyne, PA: Clinical and Laboratory Standards Institute. 2020.
- Sailaja B, Singh A. Klebsiella pneumoniae: Prevalence of ESBL Producing Clinical Isolates and their Antimicrobial Susceptibility Pattern in a Hospital Setting. Int J Curr Microbiol App Sci. 2019;8(7):2794-2799.
Crossref - Papp-Wallace KM, Endimiani A, Taracila MA, Bonomo RR. Carbapenems: Past, Present, and Future. Antimicrob Agents Chemother. 2011;55(11):4943-4960.
Crossref - Anderson K, Lonsway D, Rasheed J, et al. Evaluation of methods to identify the Klebsiella pneumoniae carbapenemase in Enterobacteriaceae. J Clin Microbiol. 2007;45(8):2723-2725.
Crossref - Tzouvelekis L, Markogiannakis A, Psichogiou M, Tassios P, Daikosc G. Carbapenemases in Klebsiella pneumoniae and Other Enterobacteriaceae: an Evolving Crisis of Global Dimensions. Clin Microbiol Rev. 2012;25(4):682-707.
Crossref - Baran I, Aksu N. Phenotypic and genotypic characteristics of carbapenem-resistant Enterobacteriaceae in a tertiary-level reference hospital in Turkey. Ann Clin Microbiol Antimicrob. 2016;15(1):20.
Crossref - Hamzan NI, Yean CY, Rahman RA, Hasan H, Rahman ZA. Detection of blaIMP4 and blaNDM1 harboring Klebsiella pneumoniae isolates in a university hospital in Malaysia. Emerg Health Threats J. 2015;8(1):26011.
Crossref - Mohanty S, Mittal G, Gaind R. Identification of carbapenemase-mediated resistance among Enterobacteriaceae bloodstream isolates: a molecular study from India. Indian J Med Microbiol. 2017;35(3):421-425.
Crossref - Veeraraghavan B, Shankar C, Karunasree S, Kumari S, Ravi R, Ralph R. Carbapenem resistant Klebsiella pneumoniae isolated from bloodstream infection: Indian experience. Pathogens and global health. 2017;111(5):240-246.
Crossref - Vivan AC, Rosa JF, Rizek CF, et al. Molecular characterization of carbapenem-resistant Klebsiella pneumoniae isolates from a university hospital in Brazil. J Infect Dev Ctries. 2017;11(5):379-386.
Crossref - Pourgholi L, Farhadinia H, Hosseindokht M, et al. Analysis of carbapenemases genes of carbapenem-resistant Klebsiella pneumoniae isolated from Tehran heart center. Iran J Microbiol. 2022;14(1):38-46.
Crossref - Ahmed MA, Yang Y, Yang Y, et al. Emergence of hyper virulent carbapenem-resistant Klebsiella pneumoniae coharboring a blaNDM-1-carrying virulent plasmid and a blaKPC-2-carrying plasmid in an Egyptian hospital. Msphere. 2021;6(3):e00088-21.
Crossref - Galani I, Nafplioti K, Adamou P, Karaiskos I, Giamarellou H, Souli M. Nationwide epidemiology of carbapenem resistant Klebsiella pneumoniae isolates from Greek hospitals, with regards to plazomicin and aminoglycoside resistance. BMC Infect Dis. 2019;19(1):167.
Crossref - Shankar C, Mathur P, Venkatesan M, et al. Rapidly disseminating bla OXA-232 carrying Klebsiella pneumoniae belonging to ST231 in India: multiple and varied mobile genetic elements. BMC Microbiol. 2019;19(1):137.
Crossref - Bhaskar H, Mulki S, Joshi H, Adhikary R. Molecular Characterization of Extended Spectrum â-lactamase and Carbapenemase Producing Klebsiella pneumoniae from a Tertiary Care Hospital. Indian J Crit Care Med. 2019;23(2):62-66.
Crossref - Han R, Shi Q, Wu S, et al. China Antimicrobial Surveillance Network (CHINET) Study Group. Dissemination of carbapenemases (KPC, NDM, OXA-48, IMP, and VIM) among carbapenem-resistant Enterobacteriaceae isolated from adult and children patients in China. Front Cell Infect Microbiol. 2020;10:314.
Crossref - Lau MY, Teng FE, Chua KH, et al. Molecular characterization of carbapenem resistant Klebsiella pneumoniae in Malaysia hospital. Pathogens. 2021;10(3):279.
Crossref - Alizadeh H, Khodavandi A, Alizadeh F, Bahador N. Molecular characteristics of carbapenem-resistant Klebsiella pneumoniae isolates producing blaVIM, blaNDM, and blaIMP in clinical centers in Isfahan, Iran. Jundishapur J Microbiol. 2021;14(2):e114473.
Crossref - Li J, Huang Z, Tang M, et al. Clonal dissemination of multiple carbapenemase genes in carbapenem-resistant enterobacterales mediated by multiple plasmids in China. Infect Drug Resist. 2021;19:3287-95.
- Kazi M, Drego L, Nikam C, et al. Molecular characterization of carbapenem-resistant Enterobacteriaceae at a tertiary care laboratory in Mumbai. Eur J Clin Microbiol Infect Dis. 2015;34(3):467-72.
Crossref - Bhatia M, Shamanna V, Nagaraj G, et al. Molecular characterisation of carbapenem-resistant Klebsiella pneumoniae clinical isolates: preliminary experience from a tertiary care teaching hospital in the Himalayas. Trans R Soc Trop Med Hyg. 2022;116(7):655-662.
Crossref - Bhatt P, Tandel K, Das NK, Grover N, Ranjan P, Rathi KR. Phenotypic detection and molecular characterization of carbapenem-resistant Enterobacteriaceae at a tertiary care center. J Mar Med Soc. 20221;24(3):40-46.
Crossref
© The Author(s) 2023. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.