ISSN: 0973-7510
E-ISSN: 2581-690X
Human hair dandruff (HHD) is a common unwanted scalp disorder that is prevalent to most human populations all over the world. This study was designed to isolate and characterize pathogens that are responsible for HHD as well as the evaluation of their biological control technique. Isolated bacteria were characterized by different biochemical tests and molecular identification methods. Here, disc diffusion methods were used to determine antibiotic and antibacterial activity against isolated bacteria. The isolated bacterial colonies were found to be Gram-positive, small, round-shaped, and purple. PCR amplification was done using 27F and 1492R primer pairs. A BlastN search of a sequenced 1465 bp region of 16S rRNA in NCBI GenBank revealed approximately 99% genome similarity with Staphylococcus aureus. The sequence was deposited in GenBank (Accession No. MH603394). In the antibiotic sensitivity test, Kanamycin showed the highest 31.0±0.5 mm diameter zone of inhibition (DZI) against the isolated bacteria. Moreover, as a plant-derived compound, the Methanol extract of Allium sativum revealed the highest, 15.0±0.5 mm DZI. The present study would give a promising direction of identification and control of this pathogen biologically.
Human hair dandruff, Molecular detection, Biological control, Staphylococcus aureus.
Human hair dandruff (HHD) is a common disorder of the skin that affects the scalp and creates an unfavorable condition for most of the people over the world1. Dandruff is associated with irritating scalp without swelling and its pathogenesis is composed of numerous intrinsic and environmental factors2. Dandruff can contribute to several causes, including dry skin, seborrhea dermatitis, inadequate washing or scrubbing, too often shampoos, scalp psoriasis, eczema, hair-care or yeast-like fungus sensitivity. Scaling and sometimes mild itching are symptoms. Seborrheic dermatitis is a more serious form of the disease, causing skin inflammation2. The severity of dandruff varies between mild and severe, where men have higher prevalence and severity3.
Bacteria are more critical than fungi to the formation of dandruff, mostly Staphylococcus and Propionibacteria4. Xue et al. 2016 reported, Staphylococcus bacterial species are more responsible for the formation of dandruff rather than fungi5. Staphylococcus aureus is gram-positive and round-shaped bacteria which are a component of the firmicutes and normal body fluids, mostly seen in the nose, breathing tract and the skin6-7. Evolution of antibiotic-resistant strains of S. aureus, for example, methicillin-resistant S. aureus (MRSA) that is a global clinical problem4. In previously reported studies, staphylococci bacteria are responsible for dandruff pathogenesis. Leong et al., 2019 reported a way to increase the capability of treatments against HDD caused by Staphylococcus8.
To address this global issue, plants are considered to be a good source of traditional medicine, as are both bioactive and new therapeutic compounds. The plants are known since ancient times and widely accepted as a crucial source of traditional medicinal compounds for specific diseases diagnosis9. There is no previously reported research where the antibacterial activity of plant extracts against human hair dandruff is discussed. We planned to treat this unfavorable scalp disorder for the first time through globally available medicinal plants.
Therefore, this study focused on the investigation of bacterial isolation from a healthy volunteer with dandruff infected hair. And to characterize the pathogen in the aspect of both biochemical and molecular approaches and to characterize the antibacterial activities of plant extracts to control this unpleasant HHD.
Hair samples collection
In present study, disease-associated hair samples were obtained from a healthy volunteer having a bad experience of this unpleasant scalp disorder who is not a member of our research group and willing to dedicate his unwanted hair scalp infected with dandruff as our research sample. Ethical clearance of the study was approved by the Director of the Institute of Biological Science, University of Rajshahi, Rajshahi-6205, Bangladesh (Approval no. IBSC.EC.5.6.18-00122).
Isolation and culture of bacteria
The hair sample collected in a sterilized zipper bag, and then hairs were taken in a conical flask that contained 100 ml distilled water. The flasks were shaken in a rotatory shaker for 30 minutes. One milliliter of water was taken into LB liquid medium by using a sterile pipette and incubated to grow bacteria at 37°C in a shaker overnight. On the next day, after incubation, a sterile loop was used to streak the bacteria onto a solid LB agar medium. The plated bacteria were cultured and incubated overnight, 16 hours, at 37°C.
The pure culture was obtained through the streak-plate method. Finally, the plate was incubated overnight at 37°C. There were many single colonies found on the plates. To obtain the desired microorganism, sub-culturing was done, and the liquid medium was prepared for every single colony. The single colonies were taken by a loop of inoculation needle and touched on LB liquid medium on the laminar flow bench. Then it was incubated for 16-18h at 37°C temperatures.
A loop of an inoculation needle was taken into the single colonies and touched on LB liquid media. It was then incubated at 37°C temperatures for 16-18 hours.
Biochemical characterization
The morphological and biochemical characteristics of the isolated bacteria were done. Bacteriological analysis was performed using selective media10-13. After 12 to 16 hours of growth in the LB Agar plate at 37°C, the morphology of the colony, size, shape, color, and growth patterns were recorded. Light microscopy was used to observe cell size. A series of biochemical tests namely, Potassium hydroxide, H2S production, Indole formation, Motility, Catalase, Simmon citrate, Kovac oxidase, Methyl red, MacConkey agar, Kligler Iron Agar(KIA), Triple Sugar Iron (TSI) and Urease tests were performed to characterize the isolated bacteria based on Bergey’s Systematic Bacteriology Manual guidelines14.
Molecular characterization
Extraction of genomic DNA
Genomic DNA was extracted from bacterial cultures by heat lysis and selective precipitation of cell debris and polysaccharides with Maxwell Blood DNA Kit (Model: AS1010) Promega, USA. The extracted DNA was re-suspended in TAE buffer (Promega, USA) and quantified using a NanoDrop Spectrophotometer (Model: ND2000, Origin: Thermo Scientific, USA), and then run 1% agarose gel electrophoresis to determine the size of the DNA.
PCR analysis
Amplification of the 16S rRNA gene was performed by conventional PCR technique using the universal primers: 27F forward primer 5’- TGGTAGTCCACGCCCTAAAC-3’ and 1492R reverse primer 5’-GACGGGCGGTGTGTRCA-3’15-16 in a 25µL reaction volume, containing nuclease free ddH2O 15μL, dNTP mix 1.0μL, forward primer 1.0μL, reverse primer 1.0μL, DNA template 1.5μL, MgCl2 2.5μL, Taq buffer B 2.5μL, and Taq polymerase 0.5μL. The PCR was done by initial denaturation at 95°C for 5 min; 35 cycles of denaturation at 95°C for 40s, annealing at 65°C for 1 min, extension at 72°C for 2 min and the final extension at 72°C for 10 min, followed by cooling at 4°C until the sample was taken out. Gel electrophoresis was used to visualize the amplified PCR product.
Antibiotic sensitivity test
A moderate disc diffusion method was applied to measure antibiotic susceptibility17. In this research work, to observe the sensitivity pattern against the isolated bacteria, 16 different antibiotics were selected. These isolated bacteria were grown overnight in a nutrient broth at 37°C in a shaker with 120 rpm for the antibiotic sensitivity test. Then, 1 ml (CFU 1X106/ml) of the overnight culture of bacteria was transferred and gently spread on the nutrient agar plate and dried. Different commercially available antibiotic disks namely, Ampicillin 10µg, Neomycin 30µg, Doxycycline 30µg, Kanamycin 30µg, Erythromycin 15µg, Tetracycline 30µg, Cefotaxime 30µg, Oxytetracycline 30µg, Azithromycin 15µg, Oxytocin 30µg, Penicillin10µg, Nalitic acid 30µg, Amoxycillin 10µg, Vancomycin 30µg, Cefixime 5µg, and Rifampicin 5µg per disc were placed over the respective plates (1 disk per plate). And then incubated at 37°C for 16 hours.
Antibacterial activity of some plant extracts
Ten diverse plant varieties such as Garlic, Onion, Hog plum, Neem, Fig tree, Cassia, Pathor Kuchi, Bitter Gourd, Ginger, and Sunflower were obtained from various locations of Rajshahi University premises. Different plant parts were used as a potent source of plant extracts, described details in Table 1. They were carefully rinsed to clean off, air-dried in sunlight for the three days (8 h/ day). And then kept in the shade in an aerated place till drying up completely, and they were pulverized to a fine powder9,18. The prepared powder was drenched in Methanol solvents followed by earlier reported an experiment with slight modification (powder to solvent ratio, 1:10, w/v), and incubated for 24 hours at 25°C with shaking at 160 rpm. Then, the filtrates from the filtration through nylon membrane clothes were dried at 37°C19. Finally, the dried extracts were dissolved in Phosphate Buffered Saline (PBS) bringing the total volume of 500 ml.
Table (1):
Plants name and plant parts (Source of plant extract) used for antibacterial activity.
Plant name |
Scientific name |
Used plant part |
|---|---|---|
Garlic |
Allium sativum |
Bulb |
Onion |
Allium cepa |
Bulb |
Hog plum |
Spondias mombin |
Fruit |
Neem |
Azadirachta indica |
Leaf |
Fig tree |
Ficus carica |
Fruit |
Cassia |
Cassia alata |
Leaf |
Pathor Kuchi |
Bryophyllum pinnatum |
Leaf |
Bitter Gourd |
Momordica charantia |
Fruit |
Ginger |
Zingiber officinale |
Root |
Sunflower |
Helianthus annuus |
Flower |
The agar disc diffusion method screened the antibacterial activity of the ten plant extracts20. A suspension of the inoculum was swabbed uniformly on the solidified 20 mL LB agar. The inoculum was allowed to dry for 5 min. Aliquot of 10μL, 20μL and 30μL from liquids crude extract of each plant (500mg/mL) was soaked into each 6 mm Whatman paper discs on the cultured medium and kept in the bench for 1h for proper diffusion and after that incubated for 24h at 37oC. The antibacterial activity of the test specimens was evaluated by measuring DZI in millimeter (mm) with a transparent scale after incubation. If the DZI is less than 10mm, it is considered as resistance. If the DZI is greater than 10mm but less than 15mm, it is considered as intermediately resistant.
Statistical analysis
For the consistency of results and statistical purposes, all the above-mentioned investigations in this research were carried out three times. The results were analyzed statistically by one-way ANOVA ( Analysis of Variances) as mean and standard error (Mean±SE) using Microsoft Excel 2013 version. For the statistical analysis, P<0.05 was considered significant.
Isolation of pure culture
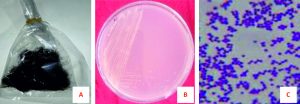
After repeated microbial culture of the isolated pathogen in suitable medium and conditions described earlier, pure culture was found. The isolated colony was creamy white (Fig.1A). The colonies were small to medium, smooth, convex, and mucoid in size and shape. The colony morphology was identified by microscopic and visual observation.
Fig. 1. Collection of hair samples, isolation of bacteria and gram staining of microorganism. (A) Hair sample (B) Isolated bacterial colonies and (C) Gram-positive bacteria
Biochemical characterization
After isolation of the pure culture, various biochemical tests, including gram-staining (noted below) was done to characterize the isolated pathogen precisely. Isolated bacteria confirmed as gram-positive both in gram staining reaction and KOH solubility test (Fig. 1B). In Sulfide Indole Motility (SIM) test, no gas was produced, no indole ring was found, and motility was observed in media (Fig. 1C). In the case of the Catalase test, bubbles (oxygen gas) were produced immediately after inoculating the bacteria with H2O2 (hydrogen peroxide), indicated catalase positive. The Simmon’s Citrate test showed dark-blue (Prussian-blue) color in the inoculating bacterial medium and the bacteria had a positive effect on the citrate medium. Isolated bacteria did not yield a purple color during the Kovac oxidase test. Therefore, it showed a negative result in the Kovac oxidase test. It was less acidic and provided yellow color following inoculation of bacteria into Methyl Red medium so that bacteria had a negative effect on methyl red. For the KIA test, it produced a yellow color on the medium following inoculation of isolated bacteria and showed positive results. For the Urease test and TSI test, negative and positive results were observed respectively by the isolated bacteria. Bacteria produced pink color rounding the colony in MacConkey agar, confirming they are lactose non-fermenting (Table 2).
Table (2):
Biochemical characteristics of the isolated bacteria.
Test |
Result |
Optimization |
Remarks |
|---|---|---|---|
Gram staining |
+ve |
Small round shaped, purple color colony |
Isolated bacteria were gram-positive |
Potassium hydroxide |
+ve |
Bacterial smear becomes viscous, sticky mess |
Isolated bacteria were gram-positive |
H2S production |
-ve |
Absence of blackening along the line of inoculation |
Bacteria were positive to H2S production |
Indole formation |
-ve |
No band is formed of the medium |
A negative indole test for isolated bacteria |
Motility |
-ve |
No growth of bacteria is observed in SIM media |
Non-motility was recorded for isolated bacteria |
Catalase |
+ve |
Copious bubbles produced |
Bacteria were able to produce catalase enzyme |
Simmon citrate |
+ve |
Production color from green to Prussian blue |
Isolated bacteria can utilize citrate |
Kovac oxidase |
-ve |
No color formation within 5-10 seconds after inoculation |
Isolated bacteria are negative to oxidase reagent |
Methyl red |
-ve |
Yellow color |
No red color was formed in methyl red |
MacConkey agar |
+ve |
Pink color for colony |
Lactose non fermenting |
Kligler Iron Agar |
+ve |
Yellow color |
Isolated bacteria non-ferment glucose |
Triple Sugar |
+ve |
Red color in slant and yellow |
Isolated bacteria could not ferment |
Iron |
color in butt |
sugars |
|
Urease |
-ve |
No color change |
No magenta color was formed |
+ve = positive, -ve= negative
Antibiotic susceptibility assay
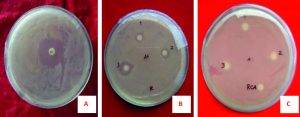
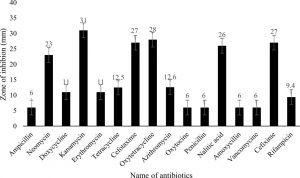
In antibiotic susceptibility assay, various types of standard discs of antibiotics against the isolated bacteria were used. The highest antibacterial activity was shown by Kanamycin (30µg/disc) with 31±0.0 mm diameter zone of inhibition (DZI) followed by Oxytetracycline (30µg/disc) with 28±0.0 mm DZI (Fig. 2A). On the contrary, Rifampicin (5µg/disc) and Doxycycline (30µg/disc) showed the lowest; 9.4±0 mm and 11±0.0mm, DZI. But Ampicillin, Oxytocin, Penicillin, and Vancomycin did not show any zone of inhibition (Fig. 3).
Fig. 2. Antimicrobial activities of drug discs against Staphylococcus aureus. (A) Kanamycin, (B) Allium sativum extract, (C) Rhizobium from Cicer arietinum.
Fig. 3. Graphical representation of antibiotics and their zone of inhibition
Legend: M±SE=mean and standard error, Resistant=<10 mm; Intermediate =10-15 mm; Susceptible=>15 mm
Antibacterial activity against isolated bacteria by some plant extracts
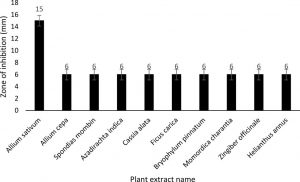
The antibiotics which cause antimicrobial resistance but have some adverse effects on human health. For this reason, the natural products with antimicrobial effects have been investigated to eliminate the use of synthetic antibiotics during the last decades. In recent decades, the use of synthetic antibiotics was replaced by natural products with antimicrobial effects. Therefore, some medicinal plants are used to inhibit the growth of the human pathogenic microorganisms. Among the 10 medicinal plants, the Methanol extract of Allium sativum showed highly significant antimicrobial activity about 15.0±0.5 mm DZI (Fig. 4). This study precisely indicates that the antibacterial activity varies with plant type and solvent type.
Fig. 4. Graphical representation of plant extracts and their zone of inhibition
Legend: Mean±SE=mean and standard error, Resistant=<10 mm; Intermediate =10-15 mm; Susceptible=>15 mm
Molecular characterization
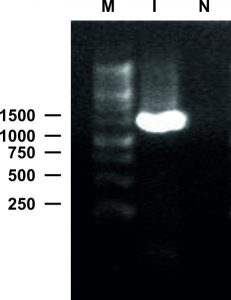
The genomic DNA of isolated bacteria was PCR amplified by one pair of universal 16s ribosomal RNA primers of the genes (27F and 1492R). The PCR amplified fragment was 1500 bp (approximately) compared to the DNA ladder of 1kb (Fig. 5).
Fig. 5 . 16s rRNA PCR profiles of 27 F and 1492 R primers, generated from bacterial genomic DNA. M: DNA ladder, I: PCR product, N: Negative Control
After sequencing, the 1465 bp nucleotide sequence was performed for a BlastN search in GenBank (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The BlastN result reported approximately 99% sequence similarity with Staphylococcus aureus strain 8 BWI 16s rDNA gene. The obtained sequence was deposited in GenBank (Accession No. MH603394).
Fig. 6. Evolutionary relationships of taxa. The isolated bacteria with its accession number is indicated by red point
Phylogenetic analysis
The sequences of Staphylococcus spp. in FASTA format were downloaded from NCBI GeneBank. The data were matched in Multiple Sequence Alignment (MSA) and constructed a phylogenetic tree (Fig. 6). The evolutionary tree was constructed using the Unweighted Pair Group Method with Arithmetic mean (UPGMA) method. The tree is drawn to scale with branch lengths that are the same units as the evolutionary distances that the phylogenetic tree is used to infer. Evolutionary steps are determined using the Composite Maximum Likelihood method. Twelve nucleotide sequences were included in the study and eliminated all positions that contain gaps and missing data. In the final dataset, there were 774 positions. Molecular Evolutionary Genetics Analysis (MEGA version 7.0) software was used to analyze the evolutionary studies21-23.
Dandruff is a disease of the skin that mainly affects the scalp2. The actual cause of this has some debates but is supposed to include a combination of environmental and genetic factors. According to a previous study reported by Xue et al. in 2016, the possibility of dandruff formation is higher due to bacteria (Staphylococcus) than that of fungi. The amount of sebum and water indicated the presence of bacteria5. In this study, the morphological, physiological, biochemical, antibiotic, antibacterial, antagonistic and molecular tests were performed after the isolation of bacterial strains for the bacterial identification. In gram staining, isolated bacteria from hair samples showed a purple color, indicating that it’s a gram-positive bacterium. Isolated bacteria showed non-motility in SIM medium and did not produce H2S and indole ring. Here, isolated bacteria showed a positive result to Simmons citrate and a negative result for the MacConkey agar test. Kligler Iron Agar (KIA) test may help to differentiate enteric gram-negative bacilli based on the fermentation of carbohydrate and production of H2S.
In this study, isolated bacteria showed a positive result to the KIA test, which means the bacteria were glucose fermenters and non-lactose fermenters. The Catalase test is used to identify organisms producing catalase enzymes. By breakdown into water and oxygen gasses, this enzyme detoxifies hydrogen peroxide. The results of bubbles produced during the production of oxygen gas indicate the Catalase test positive. This finding indicates the similarity of the previously reported experiment24. The Methyl red test is used to identify the stable acidic end product that confirms mixed acid-fermenting bacteria. In this test, isolated bacteria showed a negative result with a yellow color. Urease test measure the capability of a microorganism to breakdown urea by producing urease enzyme. Isolated bacteria showed a negative result in the present study. There was a similar result reported earlier25.
In the antibiotic susceptibility test, 16 antibiotics were used to perform the antibacterial activity of an antibiotic against bacterial isolates responsible for dandruff, using Kirby-Bauer antibiotics (KB test) or discuss susceptibility to diffusion antibiotics. An antibiotic sensitivity test was helpful in finding out the control measures of this disease. The zone of inhibition on a plate identified the sensitivity pattern of the isolated bacteria against the different types of antibiotics26. This result showed that isolated bacteria were very sensitive to Kanamycin, Oxytetracycline, Cefotaxime, Cefixime, Nalitic Acid, and Neomycin.
Antimicrobial activities against isolated bacterial species were evaluated in vitro by ten medicinal plants used as extracted material. Isolated bacteria were intermediately resistant to Allium sativum and the increase of that compound increased their sensitivity. These are similar to the findings of some previous studies26-27.
In the present study, we find out that the bacterial strain is responsible for the dandruff disease of human hair. And molecular identification of bacteria creates an opportunity to identify this pathogen using specific primers. It also provides information for future research to know the facts behind this bacterial pathogenicity.
Human hair dandruff (HDD) is a common, unpleasant scalp disorder that is one of the major concerns for the health-conscious people over the world. In our recent work, we isolated dandruff, causing pathogens from a healthy volunteer, experimented with some major biochemical and molecular characterization to control this disease using some globally available medicinal plants. In the Biochemical test, we proved that isolated bacteria as gram-positive and non-motile, glucose fermented, but lactose non-fermented. In molecular characterization, it confirms the isolated bacterial strain is Staphylococcus aureus that leads to identifying the associated genes or functional group for dandruff pathogenesis. In the antimicrobial test, Staphylococcus aureus shows the highest sensitivity to Allium sativum among all the plant extracts that recommend a new prospect for developing an anti-dandruff compound. Although there were some limitations as we isolated pathogens from a single volunteer, the present study would be helpful for further direction of identification and control of this pathogen with biologically active compounds.
Application of research
Disease identification, characterization of the pathogen and management of the above-mentioned disease will increase the quality and quantity of human hair.
ACKNOWLEDGMENTS
The authors wish to thank Professor Joarder DNA and Chromosome Research Lab, Dept. of Genetic Engineering and Biotechnology, University of Rajshahi, Bangladesh for providing financial support and other facilities during the whole research work.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
MAM, MRA, SMZH, and MFH designed the experiments, developed the methodology and prepare the manuscript. MAM, MRA, MFH, and BS collected the data and carried out the analysis. MRA, MFH, MAI, and MAI assisted with data analysis and manuscript preparation. Finally, MAM, MRA, SMZH, MAI, and MFH carried out manuscript correction according to the journal’s comment.
FUNDING
None.
ETHICS STATEMENT
Prior to the commencement of the study, the ethical clearance of the study was approved by the Director of the Institute of Biological Sciences (Approval no. IBSC.EC.5.6.18-00122) and ethical committees. In addition, specimens were collected and consent was obtained from patients.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
- Ranganathan S, Mukhopadhyay T. Dandruff: the most commercially exploited skin disease. Indian J Dermatol, 2010;55(2): 130-134.
Crossref - Borda LJ, Wikramanayake TC. Seborrheic dermatitis and dandruff: a comprehensive review. J Clin Investig Dermatol, 2015; 3(2):
Crossref - Dos Santos R, Dias-Souza M, Effectiveness of five antidandruff cosmetic formulations against planktonic cells and biofilms of dermatophytes. Saudi J Biol Sci, 2017; 24(2): 331-337.
Crossref - Lakhundi S, Zhang K. Methicillin-resistant Staphylococcus aureus: molecular characterization, evolution, and epidemiology. Clin Microbiol Rev, 2018, 31(4): e00020-18.
Crossref - Xu Z, Wang Z, Yuan C, Liu X, Yang F, Wang T et al. Dandruff is associated with the conjoined interactions between host and microorganisms. Sci. Rep, 2016; 6: 24877.
Crossref - Tong SYC, Davis JS, Eichenberger E, Holland TL, Fowler VG, Jr. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev, 2015; 28(3): 603–661
Crossref - Taylor T A, Unakal C G. Staphylococcus aureus[M]//StatPearls [Internet]. StatPearls Publishing, 2017.
- Leong C, Schmid B, Buttafuoco, A, Glatz M & Bosshard PP. In vitro efficacy of antifungal agents alone and in shampoo formulation against dandruff-associated Malassezia spp. and Staphylococcus spp. International Journal of Cosmetic Science, 2019; 41(3): 221-227.
Crossref - Parameswari P, Devika R, Vijayaraghavan P. In vitro anti-inflammatory and antimicrobial potential of leaf extract from Artemisia nilagirica (Clarke) Pamp. Saudi J Biol Sci, 2019; 26(3): 460-463.
Crossref - Prescott LM, Harley JP, Klein DA. Microbiology, 5th eds. McGraw-Hill, New York, 2002; 1014.
- Sherman N, Cappuccino JG. Microbiology: a laboratory manual. 6th eds, 2005; 81: 265-267.
- Ali MR, Hasan MF, Lia RS, Akter A, Sumi MSE, et al. Isolation and characterization of a canker disease causing pathogen from Citrus aurantifolia and evaluation of its biological control measure. J. Entomol. Zool. Stud, 2017; 5(6): 1526-1532.
- Hasan SZ, Hossain F, Zaoti ZF, Hasan F, Islam A, Khalekuzzaman, Sikdar B. PCR amplification of DNA sequence related to the hrpD gene of Xanthomonas cucurbitae in leaf spot disease of pumpkin and their antagonism by soil bacteria. Arch. Phytopathol, 2018; 51(5-6):252-66.
Crossref - Bergey DH, Holt JG, Noel RK. Bergey’s Manual of Systematic Bacteriology 9th Edn, 1994; 1: 1935-2045.
- Zaoti ZF, Hasan SZ, Hossain MF, Hasan MF et al. Biochemical and Molecular Characterization of Bacterial Wilt Disease of Banana and Evaluation of their Antibiotic Sensitivity. Microbiology Research Journal International, 2017; 22(6): 1-10, 2017.
Crossref - Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young, AC, Bouffard GG, Blakesley RW, Murray PR, Green ED and Turner ML. Topographical and temporal diversity of the human skin microbiome. Science, 2009; 324(5931): 1190-1192.
Crossref - Bauer A, Kirby W, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol, 1966; 45(4): 493-496.
Crossref - Hindi N Chabuck Z. Antimicrobial activity of different aqueous lemon extracts. J appl pharm Sci, 2013; 3: 74.
- Tchinda CF, Voukeng IK, Beng VP, Kuete V. Antibacterial activities of the methanol extracts of Albizia adianthifolia, Alchornea laxiflora, Laportea ovalifolia and three other Cameroonian plants against multi-drug resistant Gram-negative bacteria. Saudi J Biol Sci, 2017; 24(4): 950-955.
Crossref - Hasan MF, Sikdar B. Screening of antimicrobial, cytotoxic and pesticidal activities of Coccinia grandis (L.) Voigt. J. Microbiol. Biotech. Food Sci, 2016; 5(6): 584-588.
Crossref - Sneath PHA Sokal RR. Numerical Taxonomy. Freeman, San Francisco, 1973.
- Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proceedings of the National Academy of Sciences (USA), 2004; 101: 11030-11035.
Crossref - Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol, 2016; 33:1870-1874.
Crossref - Laxminarayan R, Duse A, Wattal C, Zaidi A K, Wertheim HF, et al. Antibiotic resistance the need for global solutions. Lancet Infect Dis, 2013; 13(12): 1057-1098.
Crossref - Islam MA, Mazumdar RM, Islam S, Alam MJ, Urmee SA. Isolation, Identification and In-Vitro Antibiotic Sensitivity Pattern of Citrus Canker Causing Organism Xanthomonas axonopodis. Advancements in Life Sciences, 2014; 1(4): 215-222.
- Jorgesen J Turnidge J. Susceptibility Test Methods: Dilution and Disk Diffusion Method. Manual of Clinical Microbiology, 2015; 1253-1273.
Crossref - Verma V, Singh R, Tiwari RK, Srivastava N, Verma A. Antibacterial activity of extracts of Citrus, Allium & Punica against food borne spoilage. Asian Journal of Plant Science and Research, 2012;2(4): 503-509.
© The Author(s) 2020. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.