ISSN: 0973-7510
E-ISSN: 2581-690X
There is a dynamic status carrying on continuously between the infectious bacterial agents and their surroundings. The main target of bacteria is to survive and to develop different mechanisms to overcome the hostile environment and so indirectly related to the public health hazards. One of those methods is to attain resistance genes against wide array of antimicrobial agents along to variant kinds of mineral elements presented by different concentrations. The whole genome sequence of 13 Staphylococcus (Staph) aureus isolates had been obtained and annotated. Visualization of the whole bacterial chromosome would give the chance to mine different kind of bacterial genes, and this study was concerned to look for the resistance genes against the zinc, cobalt and cadmium ions (czc). The number of resistant genes against the zinc, cobalt and cadmium varied between different isolates ranging from 2 to 4 alleles per each genome. Furthermore, the phylogenetic analysis revealed an extraordinary grade of polymorphism between different alleles within the same genome or among different ones. Conclusion: there were variable resistant genes against cobalt (Co), zinc (Zn), and cadmium (Cd) developed by the Staphylococcus aureus isolated from sub-clinical cases of mastitis.
Staph aureus, resistance Genes, cobalt, zinc, cadmium, Czc, genome sequence.
Microorganisms need certain trace elements by small concentrations such as cobalt, zinc, copper, and nickel which playing a role in different kinds of metabolic functions as coenzymes or cofactors1,2,3. This could be toxic if they exceeded the normal physiological level. Nowadays owing to the substantial contamination and the pollution of surrounding environment by excess amount of these trace elements, the bacteria have evolved wide array of mechanisms to regulate the influx and efflux processes to preserve the relatively balanced intracellular level of the heavy metallic ions1,4,5. These mechanisms either transfer the metallic ions out of cells or detoxify them so that these bacterial agents can survive in an environment containing high levels of toxic metals. However, there were different mechanisms of resistance according to each heavy metal. The genes which coding heavy metals’ resistances are placed on the microbial plasmid, or the chromosome, or both are involved6. Staphylococcal strains lacking plasmids express resistance to zinc (Zn) and cobalt (Co) that implies a plasmid-self-determining chromosomal might encrypt resistance. Conversely, operons coding Zn, Co, and cadmium (Cd) resistance found in the Alcaligenes eutrophus7 and Zn in Escherichia coli 8. At this point, we state the sequencing and inherited analysis of resistant genes on the bacterial chromosome that codes for Zn, Co, and Cd ions in Staphylococcus aureus.
Milk samples
A total of 220 samples of raw milk were obtained from the dairy cows in Qena Governorate, Egypt (120 samples) and from the dairy she-camels in Al-Shalateen area, Red Sea Governorate (100 samples). On application of clinical examination, the animals were obviously healthy showing nothing of systemic or local infection; in addition, the udder looks normal without any cardinal signs of inflammation. Samples were directly exposed to examination in 24 hours.
Isolation and culturing of Staph aureus
Preparation of Baired-Parker media was done according to previous study9. About 3 to 4 typical and large colonies were picked up to be immersed in the stock of glycerol and kept instantly at – 70 to – 80 °C until molecularly studied10.
Whole genome sequencing
The microbial DNA of 13 Staph aureus either resistant strains (MRSA) or susceptible (MSSA) were isolated, and the whole genome sequence was obtained from those extracted DNA by using the shotgun sequencing (Sanger institute UK) that divided the DNA into several contigs via several bioinformatics software’s, and the sequence of bacterial chromosome can be obtained, aligned for evaluation.
Sequence analysis and statistics
The sequence of the whole bacterial genome was manipulated by using the Artemis; “a free genome browser and annotation tool”, which lets visualization of structure’s sequence, next generation information and the outcomes of investigations within the context of the sequence, and its six-frame translation11. Alignments, translations, and comparisons of the sequence were done using the BIOEDIT (Version 7.0.9.0, Hall, T.A, 1999). The BLAST algorithm used to explore the NCBI GenBank databases for homologous sequences (http://www.ncbi.nlm.hih.gov/).
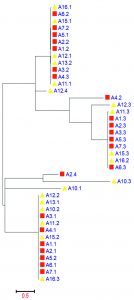
Neighbor-joining trees12 were constructed on the basis of genetic distances, as estimated by Kimura’s method13; two-parameter method, using MEGA-514, (http://www.megasoftware.net). The trustworthiness of the trees was assessed by bootstrap confidence values15 and 500 bootstrap replications were used. Zn, Co and Cd resistant genes sequences which extracted from the bacterial genomes of local isolates were used to construct the neighbor-joining tree (Table 1 and Fig. 1).
Table (1):
Showed czc gene distribution in Staphylococcus aureus isolates.
No. |
Code |
Species |
MRSA/MSSA |
czc resistance alleles |
|---|---|---|---|---|
1 |
A.1 |
cattle |
MRSA |
3 |
2 |
A.2 |
cattle |
MRSA |
4 |
3 |
A.3 |
cattle |
MRSA |
3 |
4 |
A.4 |
cattle |
MRSA |
3 |
5 |
A.5 |
cattle |
MRSA |
3 |
6 |
A.6 |
cattle |
MRSA |
3 |
7 |
A.7 |
cattle |
MRSA |
3 |
8 |
A.10 |
cattle |
MSSA |
3 |
9 |
A.11 |
camel |
MSSA |
3 |
10 |
A.12 |
camel |
MSSA |
4 |
11 |
A.13 |
camel |
MSSA |
2 |
12 |
A.15 |
camel |
MSSA |
3 |
13 |
A.16 |
cattle |
MSSA |
3 |
The entire bacterial chromosome sequencing of the local isolates of Staph aureus possibly will be annotated and pictured by RAST (Rapid Annotation Subsystem Technology) (http://rast.nmpdr.org/) and Artemis11. The resistant genes of Zn, Co and Cd were extracted and the numbers of different alleles found in each bacterial isolate were shown in Table I.
Multiple sequence alignment
The Zn, Co and Cd resistant genes (czc) were aligned and translated to their corresponding amino acid sequences; this analysis revealed that all czc genes from diverse isolates of Staph aureus either MRSA or MSSA were aligned into 4 main groups that more similar to each other. The first 3 groups, each group formed identical one except the fourth group were identical to each other. The first group was composed of the following sequences A1.1, A5.2, A2.1, A7.1, A15.2, A11.2, A6.1, A16.3, A12.2, A13.1, A4.1, A3.1 and A10.2. The second was A7.2, A5.1, A15.1, A6.2, A2.2, A1.2, A16.1, A3.2, A4.3, A12.1, A13.2 and A11.1. The third group was A11.3, A1.3, A5.3, A6.3, A7.3, A15.3, A3.3, A2.3 and A16.2. The fourth group was A12.3, A4.2, A12.4, A10.3, A2.4 and A10.1 (Fig. 2).
Table (2):
Results from Tajima’s Neutrality Test.
m |
S |
ps |
Θ |
π |
D |
|---|---|---|---|---|---|
40 |
223 |
0.991111 |
0.233008 |
0.536370 |
4.842220 |
Abbreviations: m= sequences number, n= number of total sites, S= segregation sites number, ps= S/n, Θ= ps/a1, π= diversity of nucleotides, and D= Tajima test statistic.
Neutrality test
Forty nucleotide sequences were involved in the analysis encompassing codon sites as “1st +2nd+3rd+Noncoding”. The spots comprising gaps or omitted data detected were removed. Nearby, an entire of 225 positions was in the ultimate dataset. Evolutionary evaluation was showed in MEGA6 in Table 1.
Maximum Estimate of Gamma Parameter Likelihood for Site Rates
The estimated value of the shape parameter for the “discrete Gamma Distribution” is 16.2117. Substitution pattern and rates were estimated under the Tamura-Nei 16 model (+G). A “discrete Gamma distribution” was used to show evolutionary rate changes among sites (5 categories, [+G]). Mean evolutionary rates in those categories were 0.68, 0.86, 0.98, 1.12, 1.37 substitutions per site. The nucleotide frequencies are A = 31.08 %, T/U = 35.63 %, C = 16.06 %, and G = 17.23 %. For estimating ML values, a tree topology was automatically computed. The maximum Log likelihood for this computation was -2685.629.
Phylogenetic analysis
Evolutionary status was concluded by using the “Maximum Likelihood method” grounded on the “Tamura-Nei model”. The tree with the maximum log likelihood (-2686.5090) is revealed. Initial tree(s) for the experimental search were gained by applying “Neighbor-Join and BioNJ algorithms” to a matrix of pairwise distances assessed using the “Maximum Composite Likelihood (MCL) approach”, and then selecting the topology with higher log likelihood value. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site, and those the evolutionary analyses were also conducted in the MEGA6. By evaluating the tree of the czc phylogenetic resistance genes and their different alleles, it showed that the czc alleles in the variant local isolates of Staph aureus were divided into 2 clusters, the first cluster was subdivided into 2 main subgroups. The same is applied to the second cluster. The red square is used to indicate the MRSA isolates, while the yellow triangle was used for the MSSA isolates. There is no clear demarcation between the czc alleles of either MRSA and MSSA, but each cluster contained both isolates of MRSA and MSSA. The czc allele (4) were unique and it was found in only two isolates (A2 and A12) (Fig. 1).
Dairy farm industry is suffering from many obstacles limiting their ability to produce a sufficient and clean amount of milk enough for the local consumptions especially in developing countries such as Egypt. Due to the mass use of antibacterial drugs and the high levels of contaminations by the heavy mineral elements in the surrounding environment enabled those bacteria to develop and acquire certain kind of resistant genes. The czc resistant genes against the Zn, Co and Cd were found in all isolates of Staph aureus but with variant numbers, the maximum number of czc alleles was 4 alleles in A2 and A12 isolates and the minimum number was two alleles in A13.
The czc gene was detected in all isolates of Staph aureus, as MRSA or MSSA, either. Those results came in agreement with Nair et al.17. The continuous usage of trace elements such as zinc for treatment or the routinely used disinfectants may selectively exert pressure on MRSA in the commercial herds, and the means by which such selective advantage leads to persistence of MRSA within-herd changes is still unclear, but it could be due to a combination of increased host susceptibility and increased survivability in the environment to colonization18,19. The MRSA could be derived from the early MRSA strain according to the similarity in characteristics as attributed to the clonal theory. The stability and poor transferability of methicillin resistance and similarity detected in the location of certain antibiotic resistances, included the plasmids for resistance to tetracycline, metal ions and for the beta-lactamase production20, 21. The presence of certain concentrations of resistance characters such as mineral elements suggested the buildup of resistance on bacterial chromosome by the insertion of sequence element (IS431) in the locus rnec could present a recombining site for resistances22.
This study was concerned with the search for resistance genes against Zn, Co and Cd, the czc gene was found in all isolates of Staph aureus. The number of czc alleles varied from genome to another, the minimum number was 2 alleles and the maximum number was 4 alleles. By application of the phylogenetic analysis, there was great variation in the nucleotide sequences between the different alleles of czc genes belonging to different genomes.
ACKNOWLEDGMENTS
The authors are grateful to the Deanship of Scientific Research at King Khalid University for support, and to Prof. Mark Holmes, University of Cambridge, UK, for the collaboration during the recruitment process and provided surveillance data.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors. We just collected the animal samples.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
- Nies DH, Silver S. Cloning and expression of plasmid genes encoding resistances to chromate and cobalt in Alcaligeneseutrophus. J. Bacteriol., 1989; 171: 5065-5070.
Crossref - Sambrook J Green MR. Molecular cloning: A laboratory manual, 2012; 4th ed. www.cshlpress.com/pdf/sample/2013/MC4/MC4FM.pdf
- Ali OA. Molecular epidemiology based on SPA genotyping of Staphylococcus aureus Isolated from cattle and camels in Egypt. Alex. J. Vet. Sci., 2016; 48: 62-68.
Crossref - Tabor S, Richardson CC. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc. Nat. Acad. Sci. USA (PNAS), 1985; 82: 1074-1078.
Crossref - Tsai KL, Yoon KP, Lynn AR. ATP-dependent cadmium transport by the cadA cadmium resistance determinant in everted membrane vesicles of Bacillus subtilis. J. Bacteriol., 1992; 174: 116-121.
Crossref - Van der Lelie D, Schwuchow U, Schwidetzky S, Wuertz W, Mergeay BM, Nies DH. Two component regulatory systems involved in transcriptional control of heavy metal homoeostasis in Alcaligenes eutrophus. Mol. Microbiol., 1997; 23: 493-503.
- Nies DH, Silver S. Ion efflux systems involved in bacterial metal resistances. J. Indust. Microbiol., 1995; 14: 186-199.
Crossref - Conklin DS, McMaster JA, Culbertson MR, Kung C. COT1, a gene involved in cobalt accumulation in Saccharomyces cerevisiae. Mol. Cell Biol., 1992; 12: 3678-3688.
Crossref - Vanderzant C, Splittstoesser DF. Compendium of methods for the microbiological examination of foods. Amer. Public Health Assoc., 1992; 3rd Ed: p. 20-35.
- Jones et al. Maintenance of bacteria on glass beads at – 60 °C to -76 °C. In: Kirsop and Doyle (eds.) Maintenance of microorganisms and cultures cells. Acad. Press. 1991; 2nd Ed, p. 45-50.
- Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. Artemis: Sequence visualization and annotation. Bioinformatics (Oxford England),2000; 16: 944-945.
Crossref - Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evolut.,1987; 4: 406-425.
Crossref - Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evolut.,1980; 16: 111-120.
Crossref - Kumar S, Tamura K, Jakobsen IB, Nei M. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics,2001; 17: 1244-1245.
Crossref - Felsenstein J. Phylogenies and the comparative Method. Amer. Nat., 1985; 125: 1-15.
Crossref - Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evolut.,1993; 10: 512-526.
Crossref - Nair R, Thapaliya D, Su Y, Smith TC. Resistance to zinc and cadmium in Staphylococcus aureus of human and animal origin. Infect. Cont. Hosp. Epidemiol., 2014; 35: S32-9.
Crossref - Slifierz MJ, Friendship RM, Weese JS. Methicillin-resistant Staphylococcus aureus in commercial swine herds is associated with disinfectant and zinc usage. Appl. Environ. Microbiol., 2015; 81: 2690-95.
Crossref - Ali AO, Mahmoud HYAH. ‘Epidemiological studies based on multi-locus sequence typing genotype of methicillin susceptible Staphylococcus aureus isolated from camel’s milk’. Onderstepoort J. Vet. Res., 2017; 84: a1425.
Crossref - Lacey RW. Genetic control in methicillin-resistant strains of Staphylococcus aureus. J. Med. Microbiol., 1972; 5: 497-508.
- Lacey RW, Grinsted J. Genetic analysis of methicillin-resistant strains of Staphylococcus aureus evidence for their evolution from a single clone. J. Med. Microbiol., 1973; 6: 511-526.
Crossref - Skinner S, Inglis B, Matthews PRY, Stewart PR. Mercury and tetracycline resistance genes and flanking repeats associated with methicillin resistance on the chromosome of Staphylococcus aureus. Mol. Microbiol., 1988; 2: 289-292.
Crossref
© The Author(s) 2020. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.