ISSN: 0973-7510
E-ISSN: 2581-690X
This study aimed to investigate the genetic basis for quinolones and fluoroquinolones insusceptibility in E.coli isolated from outpatients suspected with urinary tract infections (UTIs). Fifty-one nalidixic acid unsusceptible E. coli isolates were collected from adult patients with UTIs . Antimicrobial sensitivity testing was performed by disc diffusion test, and minimum inhibitory concentrations of nalidixic acid and ciprofloxacin were determined by E. test strips. PCR amplification and DNA sequencing were carried out for the detection of alterations in the quinolone resistance determining region (QRDR) of gyrA and parC genes and screening for plasmid-mediated quinolone resistance (PMQR) genes (qnr, aac(6)-ib-cr, qepA and oqxAB). Genetic analysis of the QRDR revealed amino acid substitutions in the codons 83 and 87 of the gyrA gene with or without alterations at codons 80 and 84 of the parC gene. Most of the quinolone-resistant isolates (94%) had at least one of the PMQR genes. QRDR mutations in chromosomal genes encoding gyrA and parC have the principal role in the development of quinolones and fluoroquinolnes resistance in E.coli. The accumulation of amino acid alterations in gyrA and the concurrent mutations in parC lead to the emergence of a high-level of insensitivity toward fluoroquinolones. The detected PMQR determinants are widespread in the community, and they are associated with the development of resistance to many other categories of antibiotics. This is the first report to detect oqxAB in E.coli in Egypt.
E. coli, Urinary tract infection, Egypt, oqxAB
In 1885, Theodor Escherich isolated E. coli from the faeces of neonates and described it as bacterium coli commune. It was later named Escherichia coli1. Although some strains of E. coli are significant pathogens of humans and animals, other strains are harmless commensals of the intestinal tract. The pathogenic strains are separated into two groups: those causing diseases inside the intestinal tract and others capable of infection at extra-intestinal sites as the uropathogenic E. coli (UPEC) which is the most common organism implicated in urinary tract infections (UTIs)2,3.
Quinolones are synthetic antibacterial agents with excellent action against Enterobacteriaceae4. In 1962 the first quinolone, nalidixic, acid was discovered. This detection led to the rise of a variety of quinolones. Structural modification by adding a fluorine atom at C-6 position led to the invention of fluoroquinolones which are more systemically active. The bactericidal activity of quinolones comes from the inhibition of DNA gyrase and topoisomerase IV; both are involved in DNA replication5. The extensive usage of quinolones was responsible for spreading quinolone resistance in different microorganisms6.
Quinolone resistance is caused mainly as a result of amino acid alterations in quinolone resistance determining regions (QRDRs) of DNA gyrase and topoisomerase IV. Other mechanisms include decreased membrane permeability, overexpression of efflux pumps and plasmid-mediated quinolone resistance (PMQR) genes7.
The first PMQR mechanism the qnr gene (currently qnrA1), was detected in 1998. Qnr genes protect DNA gyrase and/or topoisomerase IV from fluoroquinolone action8,9. Secondly; the aac (6’)-Ib-cr which is a variant from the aminoglycoside acetyltransferase (aac(6᾿)-Ib) enzyme. This enzyme acts by N-acetylation of the amino nitrogen on the unsubstituted piperazinyl substitute of ciprofloxacin, and norfloxacin6. The third PMQR mechanism includes active efflux pumps: the OqxAB and the QepA, which reduce sensitivity to hydrophilic fluoroquinolones such as ciprofloxacin10,11.
This study was designed to detect the mechanisms of quinolone resistance among E. coli isolates from patients with UTIs in Egypt.
Clinical Isolates
Two hundred and eighty -left over, remnants- urine samples from adult patients suspected with UTIs from outpatient sections at the clinical microbiological laboratory at Fayoum University Hospital and other private laboratories of medical analysis from September 2014 to August 2015 at Fayoum governorate, Egypt, were screened for E. coli by conventional bacterial identification tests and confirmed by Microbact Identification Test, (Oxoid, UK).
Antibiotic sensitivity testing
The isolates were subjected to antimicrobial sensitivity tests to nalidixic acid, ciprofloxacin, ampicillin, amoxicillin/clavulanate, ceftazidime, cefoxitin, cefotaxime, ceftriaxone, gentamicin, amikacin, and imipenem (Oxoid, UK) according to routine laboratory protocols of clinical and laboratory standards institute (CLSI)12. The MIC of nalidixic acid has been proposed to could be used as a phenotypic marker of insensitivity for quinolones in Gram-negative bacteria13, so that the MIC of nalidixic acid was used to detect quinolone resistance among isolates. The MICs of nalidixic acid and ciprofloxacin were determined by E.test (Liofilchem, Italy). E.coli ATCC 25922 strain was used as a negative control.
Phenotypic detection of Extended-Spectrum Beta-lactamases
The phenotypic revealing of ESBLs was proceeded by the double-disc potentiation test14. It was performed by centralizing an Amoxicillin/Clavulanic (20/10µg) disc with cefotaxime(30µg), cefpodoxime(10µg), ceftazidime(30µg) and cefepime(30µg) discs strategically placed 20 to 30 mm away on Muller Hinton agar which already had a lawn of the test organism. After overnight incubation, an extension of an inhibition zone toward the AMC disc was considered positive.
Genetic analysis of resistance mechanisms
DNA extraction
DNA was extracted by using genomic pure link extraction kits (Invitrogen, USA), and according to the manufacture̕ s extraction guidelines.
PCR of the QRDR of gyrA and parC genes
QRDR of the gyrA and parC genes of selected isolates with different ciprofloxacin MICs were amplified by PCR using specific primers (Table 1). A volume of 5µl DNA was added to the 20µl reaction mixture containing 12.5µl PCR reaction mix (Sigma, USA), 1µl of each primer and 5.5µl dH2O. Reaction mixed without DNA template served as negative controls.
Table (1):
Oligonucleotide primers used for PCR amplification and sequencing of resistance genes.
| Gene | Primer name. | Sequence (5̕ to 3̕ ) | Annealing temperature | Amplicon size (bp) | Reference |
|---|---|---|---|---|---|
| gyrA | gyrA6-F | CGACCTTGCGAGAGAAAT | 55 | 620 | 38 |
| gyrA631-R | GTTCCATCAGCCCTTCAA | ||||
| parC | parC-F | TGAATTTAGGGAAAACGCCTA | 55 | 559 | 39 |
| parC-R | GCCACTTCACGCAGGTTATG | ||||
| Aac(6’)-Ib | aac(6’)-Ib-F | TTGCGATGCTCTATGAGTGGCTA | 58 | 482 | 40 |
| aac(6’)-Ib-R | CTCGAATGCCTGGCGTGTTT | ||||
| qnrA | qnrA-F | ATTTCTCACGCCAGGATTTG | 55 | 516 | 6 |
| qnrA-R | GATCGGCAAAGGTTAGGTCA | ||||
| qnrB | qnrB– F | GATCGTGAAAGCCAGAAAGG | 55 | 469 | 6 |
| qnrB– R | ACGATGCCTGGTAGTTGTCC | ||||
| qnrS | qnrS– F | ACGACATTCGTCAACTGCAA | 55 | 417 | 6 |
| qnrS-R | TAAATTGGCACCCTGTAGGC | ||||
| qepA | qepA-F | GCAGGTCCAGCAGCGGGTAG | 60 | 199 | 10 |
| qepA-R | CTTCCTGCCCGAGTATCGTG | ||||
| oqxA | oqxA-F | CTCGGCGCGATGATGCT | 57 | 392 | 11 |
| oqxA-R | CCACTCTTCACGGGAGACGA | ||||
| oqxB | oqxB-F | TTCTCCCCCGGCGGGAAGTAC | 64 | 512 | 11 |
| oqxB-R | CTCGGCCATTTTGGCGCGTA |
PCR for detection of PMQR genes
For all PMQR genes, the following reaction mixture was used: 1μl template DNA was added to 24μl PCR master mix (Sigma, USA), which contained 12.5μl PCR reaction mix, 1μl of each primer and completed to 25μl with dH2O. Specific primers used are listed in Table 1.
Sequencing
All PCR yields were exposed to electrophoresis in 1% agarose gel having ethidium bromide 0.5 mg/L. After purification, Amplified fragments were sent to Macrogen Company (Macrogen Inc., Seoul, Korea) for sequencing. Nucleotide sequences were determined on both strands of PCR amplification products. Investigation and comparison of nucleotide sequences was performed using programs accessible at the NCBI web site (http://www.ncbi.nlm.nih.gov).
Table (2):
Results of the disc diffusion test of the Nalidixic acid resistant isolates (N=51) (CLSI, 2014).
Antibiotic |
No.of resistant isolates |
% |
|---|---|---|
Nalidixic acid (30ug). |
51 |
100.0 |
Ampicillin (10ug). |
51 |
100.0 |
Cefoperazone (75ug). |
40 |
78.4 |
Cefoxitin(30ug). |
39 |
76.5 |
Ceftazidime (30ug). |
39 |
76.5 |
Ciprofloxacin (5ug). |
37 |
72.5 |
Norfloxacin(10ug) |
37 |
72.5 |
Ofloxacin(5ug) |
37 |
72.5 |
Levofloxacin (5ug) |
37 |
72.5 |
Amoxicillin/Clavulanic (20/10ug). |
36 |
70.6 |
Ceftriaxone (30ug). |
35 |
68.6 |
Gatifloxacin(5ug) |
35 |
68.6 |
Cefotaxime (30ug). |
34 |
66.7 |
Gentamicin (10ug). |
30 |
58.8 |
Amikacin (30ug). |
14 |
27.5 |
Imipenem (10ug). |
0 |
0.0 |
Out of all uropathogens recovered (n=280), 155(55.4%) were identified as E. coli. They were serially numbered from Y1 to Y155.
Antibiotic sensitivity test results
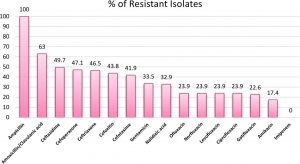
The sensitivity pattern of isolated E.coli strains is shown in Fig. 1.
Fig. 1. Antibiotic Sensitivity pattern of isolated E.coli strains from urine samples of patients with UTIs (N=155)
Fifty-one (32.9%) nalidixic acid-resistant; MIC>16µg/ml12 E. coli isolates were recorded from the isolated E.coli strains.
All of the nalidixic acid-resistant isolates were; resistant to ampicillin, sensitive to imipenem and all of them were multi drug-resistant (MDR). Thirty-seven isolates (62.5%) were sensitive to amikacin, according to the results of Ciprofloxacin MIC12, 12(23.5%) of isolates were sensitive to ciprofloxacin; MIC<1µg/ml, 2(3.9%) were intermediately resistant; MIC=1, and 37(72.5%) were resistant and highly resistant to ciprofloxacin; MIC >1. The Distribution of MIC values of ciprofloxacin is shown in Table 3.
Table (3):
The MICs of the isolates for ciprofloxacin(N=51) (CLSI, 2014).
| MIC (µg/ml) | NO. Of isolates | S/ I /R | % |
|---|---|---|---|
| ≤0.064 | 8 | S |
19.6 |
| 0.125 | 1 | S | |
| 0.5 | 1 | S | |
| 1 | 2 | I | 3.9 |
| 2 | 4 | R |
76.5
|
| 4 | 2 | R | |
| 8 | 1 | R | |
| 16 | 2 | R | |
| ≥32 | 30 | R |
MICs (µg/ml) were determined by E.test; R, resistant; I, intermediate; and S, susceptible.
Phenotypic detection of ESBLs
The double disc potentiation test showed synergy between amoxicillin/clavulanic and one or more of the following 3rd, 4th cephalosporin : cefotaxime, ceftazidime, aztreonam, and ceftriaxone in all isolates suggesting the production of ESBLs.
PCR and sequencing of gyrA and parC
Sequencing of QRDR of the gyrA and parC genes revealed mutations at the positions 83 (ser→Leu) and 87 (Asp→Asn) in gyrA and the positions 80 (Ser→Ile) and 84 (Glu→Val) in parC. At least one amino acid substitution in the gyrA gene at the codon 83 (Ser83Leu) was identified in each quinolone resistant isolate (Table 4).
Table (4):
Results of the sequencing of QRDR of selected isolates.
| Strain | MIC.CIP. | Mutations | PMQR determinant | |
|---|---|---|---|---|
| gyrA | parC | |||
| Y10 | 0.064 | S83L | ||
| Y77 | 1 | S83L | aac(6)-Ib-cr | |
| Y44 | 0.25 | S83L | ||
| Y16 | 0.5 | S83L | ||
| Y13 | 32 | S83L,D87N | aac(6)-Ib-cr | |
| Y52 | 32 | S83L,D87N | S80I | |
| Y63 | 16 | S83L,D87N | S80I | aac(6)-Ib-cr |
| Y66 | 2 | S83L,D87N | S80I | |
| Y81 | 2 | S83L,D87N | S80I | |
| Y1 | 32 | S83L,D87N | S80I,E84V | aac(6)-Ib-cr + qepA |
| Y6 | 32 | S83L,D87N | S80I,E84V | aac(6)-Ib-cr + qnrS |
| Y7 | 32 | S83L,D87N | S80I,E84V | aac(6)-Ib-cr + qnrS + qepA |
| Y15 | 32 | S83,D87N | S80I,E84V | aac(6)-Ib-cr + qnrS + oqxAB |
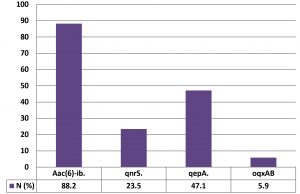
Detection of PMQR genes
At least one PMQR gene was detected in most of the nalidixic acid-resistant E.coli isolates (94%). QnrS gene was located in 12 (23.5 %) isolates. The sequencing of the qnrS gene was identical to that of the variant qnrS1. Forty-five of the 51 isolates (88.2%) were positive to aac (6)-Ib. The sequencing of the aac (6′)-Ib-cr was identical to the –cr variant. QepA gene was detected in twenty-four of the 51 isolates (47.1%). The oqxAB gene was identified in three isolates (5.9 %). This is the first report for detecting oqxAB gene in quinolone-resistant urinary isolates in Egypt. The sequencing of the oqxAB gene was identical to that of the oqxAB1 (Fig. 2).
The resistance of E. coli to fluoroquinolones which is one of the most widely used medicines for the treatment of UTIs is prevalent. Fluoroquinolone treatment is no longer effective in more than half of patients in many countries in the world15.
In the present study, 55.4% of all uropathogens were E. coli. This result agrees with that reported in previous studies in which E.coli was the leading reason of UTIs3,16,17.
Also, sensitivity pattern of the isolated E. coli strains agrees with previous studies18,19.
All of the isolated E.coli strains were resistant to ampicillin and sensitive to imipenem; this result is consistent with that reported by Mendonca et al.20. Imipenem appears to have a broader range of activity than other tested antimicrobial agents. The explanation for that is probably the fact that these are potent drugs used only in hospital settings and not as first-line therapies, so they are of limited use.
Ciprofloxacin resistance observed was higher than that described in prior studies from Egypt21. This growing in ciprofloxacin insusceptibility rate may be due to the extensive usage of quinolones, the extended use of small doses of the more effective fluoroquinolones such as ciprofloxacin, also the inappropriate use of fluoroquinolones6,22.
ESBL production was recorded for all quinolone-resistant isolates. This finding is consistent with that reported by another previous study11. Also, a study conducted in Europe reported quinolone resistance in 92% of ESBL-producing isolates, this high prevalence of quinolone-resistant ESBL-producing E. coli strains had become a significant health problem, making treatment of bacterial infections more challenging and resulting in higher morbidity and mortality rates23.
In this study, all quinolone-resistant E.coli isolates collected were MDR (Resistance to one or more antibiotics in three or more antibacterial classes)24. MDR bacterial infections are relatively common now in the community as well as the hospitals25. This High predominance of MDR isolates of E. coli has been detected in many developing countries22,26. The high incidence of MDR and ESBL production among these E.coli isolates may be attributed to many reasons. The most apparent reason is plasmids which carry the genes encoding ESBLs, and also carry the genes encoding multi-resistance to several antibiotics, including quinolones, further contributing to the spreading of resistance to several antimicrobials through inappropriate use of quinolones, this announces that these plasmid transmitted genes are clinically significant23.
Insusceptibility to quinolones and fluoroquinolones happens essentially because of amino acid substitutions in gyrA and parC genes13,27.
Sequencing of QRDR of the gyrA and parC genes revealed mutations in gyrA gene at two codons: the first is codon 83 where serine is substituted by leucine; the second is codon 87 where Asparagine is substituted by Aspartic acid. The secondary target for quinolone-resistance is the parC gene. In the current study, Point mutations at the codons 80 and 84 in parC gene were detected in many of the isolates, where Serine was substituted by Isoleucine at codon 80 and Glutamic acid is substituted by Valine at codon 84. One mutation in the gyrA gene at codon 83 was detected at least in each quinolone resistant E.coli isolate. Additional mutations in other positions in the QRDR of gyrA and parC were observed in highly quinolone and fluoroquinolone resistant isolates. The reported mutations at the QRDR in this study agree with many previous studies9,13,27,37 and prove that this is the main quinolone and fluoroquinolone resistance mechanism in E.coli and other bacterial species.
MICs of ciprofloxacin were different among E. coli isolates with the same types of amino acid alterations in gyrA and parC. This diversity may be due to other mechanisms involved in fluoroquinolone resistance such as decreased membrane permeability and overexpression of efflux pumps11.
PMQR genes play an important role in the resistance to quinolones and fluroquionolones because of their horizontal transfer and they increase the mutant prevention concentration of quinolones, thus enabling the development of mutants with a higher level of resistance to fluoroquinolones by favoring selection of amino acid substitutions in the QRDR genes8,9. PMQR genes are commonly distributed among Enterobacteriaceae28.
The first PMQR gene is Qnr. Qnr genes bind to DNA gyrase and/or topoisomerase and so they protect them from quinolones. The first detected Qnr gene in Egypt was Providencia spp. 29. In the current study, qnrs1 was the only detected qnr gene, it was detected in 23.5% of the isolates. This result agrees with previous studies from Egypt that described that qnrS as the most prevalent qnr determinant in Egypt31,32. But other Egyptian studies reported qnrB in a higher rate than the qnrS32,33. This may be due to the different bacterial strains and clinical infections included in that studies.
The second PMQR gene detected was the aac(6’)-Ib-cr, which is a variant of the gene AAC(6’)-Ib. This enzyme reduces only quinolones with piperazinyl nitrogen (ciprofloxacin and norfloxacin) activity by acetylation6. Screening for aac(6’)-Ib-cr shows a high predominance among the quinolones resistant E. coli isolates where 88.2% of the isolates were positive for it. This result agrees with that reported by Yang et al.34.
The most predominant PMQR gene found in these isolates was aac(6’)-Ib-cr. This result was expected because all the isolates were ESBL producers, and many studies have reported its prevalence among other populations of E. coli, mostly among those produce ESBL enzymes9. It was found that all qnr positive isolates were also harboring aac(6‘)-Ib-cr gene. The co-existence of the qnr and aac(6`)-Ib-cr genes on the same plasmid may be the explanation for that35.
Recently, qepA and oqxAB, multidrug efflux pumps genes, were discovered and reported as the third PMQR mechanism involved in quinolone resistance10,11. In the present study, the prevalence of qepA among the isolates (47.1%) was higher than that in a previous study from Egypt36, this high prevalence may be attributed to that most of the isolates were ciprofloxacin resistant with a high MIC.
The OqxAB efflux pump is encoded by the oqxA and oqxB genes, which are located in the same operon. In this study, the plasmid mediated oqxAB was detected in 5.9 % of the E. coli isolates. This result agrees with that reported by Yuan et al.37.
This study concluded that fluoroquinolone resistance is in growing rate in Egypt. This data supports our understanding of the molecular mechanisms of quinolone and fluoroquinolone insensitivity due to the accumulation of amino acid substitutions in the QRDR of gyrA and parC genes and PMQR determinants. Due to the improper and increased usage of wide-spectrum antibacterials and quinolones, it is essential to restrict the use of these antibiotics to be prescribed only when necessary and only when approved by a physician. The detected high prevalence of PMQR determinants through this study in Egypt, because of unrestrained quinolone use, is troublesome.Additionally, other insensitivity genes usually are harbored by these plasmids, causing multiple resistances among isolates which can complicate therapeutic management of infections and also can be transferred to other bacterial species. This study is the first report to detect plasmid mediated oqxAB in E.coli isolates in Egypt.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
ETHICS STATEMENT
This study was approved by the Research Ethical Committee at Fayoum Faculty of Medicine, Fayoum, Egypt.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript
- Todar K. “Pathogenic E. coli: Todar’s Online Textbook of Bacteriology”, 2008.
- Kaper James B, James P Nataro, Harry LT Mobley. “Pathogenic Escherichia coli.” Nat Rev Microbiol. 2004;2(2):123.
Crossref - Nicolle Lindsay E. “Uncomplicated urinary tract infection in adults including uncomplicated pyelonephritis.” Urologic Clinics. 2008;35(1):1-12.
Crossref - Soto Sara M, Joaquim Ruiz, M Carmen Mendoza, Jordi Vila. “In vitro fluoroquinolone-resistant mutants of Salmonella enterica serotype Enteritidis: analysis of mechanisms involved in resistance.” Int J Antimicrob Agents. 2003;22(5): 537-540.
Crossref - Andriole Vincent T. “The quinolones: past, present, and future.” Clin Infect Dis. 2005;41(Supp.2):S113-S119.
Crossref - Robicsek Ari, George A Jacoby, David C Hooper. “The worldwide emergence of plasmid-mediated quinolone resistance.” Lancet Infect Dis. 2006;6(10):629-640.
- Cao Xiaoli, Lina M Cavaco, Yuan Lv, et al. “Molecular characterization and antimicrobial susceptibility testing of Escherichia coli isolates from patients with urinary tract infections in 20 Chinese hospitals.” J Clin Microbiol. 2011;49(7):2496-2501.
Crossref - Martinez-Martinez, Luis, Alvaro Pascual, George A. Jacoby. “Quinolone resistance from a transferable plasmid.” The Lancet. 1998;351(9105):797-799.
Crossref - Baudry-Simner, Patricia J, Amanpreet Singh, James A Karlowsky, Daryl J Hoban, George G. Zhanel and Canadian Antimicrobial Resistance Alliance. “Mechanisms of reduced susceptibility to ciprofloxacin in Escherichia coli isolates from Canadian hospitals.” Canadian Journal of Infectious Diseases and Medical Microbiology. 2012;23(3):e60-e64.
Crossref - Yamane Kunikazu, Jun-ichi Wachino, Satowa Suzuki, Yoshichika Arakawa. “Plasmid-mediated qepA gene among Escherichia coli clinical isolates from Japan.” Antimicrob Agents Chemother. 2008;52(4):1564-1566.
Crossref - Kim Hong Bin, Minghua Wang, Chi Hye Park, Eui-Chong Kim, George A Jacoby, David C Hooper. “oqxAB encoding a multidrug efflux pump in human clinical isolates of Enterobacteriaceae.” Antimicrob Agents Chemother. 2009;53(8):3582-3584.
Crossref - Clinical and Laboratory Standards Institute, M100-S24 Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fourth Informational Supplement. CLSI: Wayne, PA, 2014.
- Ruiz Joaquim. “Mechanisms of resistance to quinolones: target alterations, decreased accumulation and DNA gyrase protection.” Antimicrob Agents Chemother. 2003;51(5):1109-1117.
Crossref - Jarlier Vincent, Marie-Helene Nicolas, Genevieve Fournier, Alain Philippon. “Extended broad-spectrum β-lactamases conferring transferable resistance to newer β-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns.” Clin Infect Dis. 1988;10(4):867-878.
Crossref - World Health Organization. Antimicrobial Resistance. Available at http://www.who.int/mediacentre/factsheets/fs194/en/ (cited 29 January 3:21 AM, 2017).
- Daoud, Ziad, Claude Afif. “Escherichia coli isolated from urinary tract infections of Lebanese patients between 2000 and 2009: epidemiology and profiles of resistance.” Chemotherapy Research and Practice,2011.
Crossref - Niranjan V and A Malini. “Antimicrobial resistance pattern in Escherichia coli causing urinary tract infection among inpatients.” Indian J Med Res.2014;139(6):945.
- Dash Sandeep Kumar, Subhankari Prasad Chakraborty, Debasis Mandal, Somenath Roy. “Isolation and characterization of multidrug-resistant uropathogenic Escherichia coli from urine sample of Urinary tract infected patients.” Int J Life Sci Pharma Res. 2012;2:25-39.
- El-Sokkary Mohamed Mohamed Adel, Eman Salama Abdelmegeed. “Antibacterial resistance pattern among Escherichia coli strains isolated from Mansoura hospitals in Egypt with a special reference to quinolones.” Afr J Microbiol Res. 2015;9(9):662-670.
Crossref - Mendonca Nuno, Joana Leitao, Vera Manageiro, Eugenia Ferreira, Manuela Canica and Antimicrobial Resistance Surveillance Program in Portugal. “Spread of extended-spectrum β-lactamase CTX-M-producing Escherichia coli clinical isolates in community and nosocomial environments in Portugal.” Antimicrob Agents Chemother. 2007;51(6):1946-1955.
Crossref - El Kholy Amani, Hadia Baseem, Geraldine S Hall, Gary W Procop, David L Longworth. “Antimicrobial resistance in Cairo, Egypt 1999–2000: a survey of five hospitals.” Antimicrob Agents Chemother. 2003;51(3):625-630.
Crossref - Ibrahim ME, NE Bilal, ME Hamid. “Increased multi-drug resistant Escherichia coli from hospitals in Khartoum state, Sudan.” Afr Health Sci. 2012;12(3):368-375.
Crossref - Kilinc Cetin, Melek Inci, Erkan Yula, et al. “Molecular Epidemiology and Risk Factors in Extended-Spectrum Beta-Lactamase Positive and Quinolone-Resistant Escherichia coli Strains Isolated from Urinary Tract Infections.” Acta Medica. 2015;31:1035.
- Magiorakos A-P, A Srinivasan, RB Carey, et al. “Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance.”Clin Microbiol Infect. 2012;18(3):268-281.
Crossref - Levy Stuart B, Bonnie Marshall. “Antibacterial resistance worldwide: causes, challenges and responses.” Nature medicine. 2004;10(12):S122-S129.
Crossref - Malhotra Ranjana, Rama Sikka, Uma Chaudhary. “Antimicrobial sensitivity pattern among clinical isolates of Escherichia coli in tertiary care centre of Northern India.” J Res Med Sci. 2016;4(2):639-642.
Crossref - Ito Carmen Antonia Sanches, Ana Cristina Gales, Maria Cristina B Tognim, PatrIcia Munerato, Libera Maria Dalla Costa. “Quinolone-resistant Escherichia coli.” Braz J Infect Dis. 2008;12(1):5-9.
Crossref - Cao X, Zhang Z, Shen H, Ning M, Chen J, Wei H & Zhang K. Genotypic characteristics of multidrug-resistant Escherichia coli isolates associated with urinary tract infections. APMIS, 2014; 122(11): 1088-1095.
Crossref - Wiegand I, N Khalaf, MHM Al-Agamy, B Wiedemann. “First detection of the transferable quinolone resistance determinant in clinical Providencia stuartii strains in Egypt.” Clin Microbiol Infect. 2004;Supplement10:64.
- El-Essawy A K, Abu Shady HM, BM El-Biaa. “The emergence of plasmid-mediated quinolone resistance genes qnrA, qnrB, and qnrS among Egyptian clinical isolates of Enterobacteriaceae.” Afr J Microbiol Res. 2011;5(30):5384-5397.
- El-Mahdy Rasha H. “Plasmid-Mediated Quinolone Resistance (PMQR) Determinants in Nosocomial Isolates of Enterobacteriaceae.” The Egyptian Journal of Medical Microbiology (EJMM). 2016;24(1).
- Kotb Dalia Nabil, Wafaa Khairy Mahdy, Mahmoud Shokry Mahmoud, Rasha MM Khairy. “Impact of co-existence of PMQR genes and QRDR mutations on fluoroquinolones resistance in Enterobacteriaceae strains isolated from community and hospital acquired UTIs.” BMC Infectious Diseases. 2019;19(1):979.
Crossref - El-Badawy Mohamed F, Wael M Tawakol, Shaymaa W El-Far, et al. “Molecular identification of aminoglycoside-modifying enzymes and plasmid-mediated quinolone resistance genes among Klebsiella pneumoniae clinical isolates recovered from Egyptian patients.” International Journal of Microbiology. 2017.
Crossref - Yang Hee Young, You Sun Nam, Hee Joo Lee. “Prevalence of plasmid-mediated quinolone resistance genes among ciprofloxacin-nonsusceptible Escherichia coli and Klebsiella pneumoniae isolated from blood cultures in Korea.” Canadian Journal of Infectious Diseases and Medical Microbiology. 2014;25(3):163-169.
Crossref - Jiang Yan, Zhihui Zhou, Ying Qian, et al. “Plasmid-mediated quinolone resistance determinants qnr and aac (6′)-Ib-cr in extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae in China.” J Antimicrob Chemother. 2008;61(5):1003-1006.
- Hassan WM, A Hashim, RAA Domany. “Plasmid-mediated quinolone resistance determinants qnr, aac (6′)-Ib-cr, and qep in ESBL-producing Escherichia coli clinical isolates from Egypt.” Indian J Med Microbiol. 2012;30(4):442-447.
Crossref - Yuan Jinyi, Xiaogang Xu, Qinglan Guo, et al. “Prevalence of the oqxAB gene complex in Klebsiella pneumoniae and Escherichia coli clinical isolates.” J Antimicrob Chemother. 2012;67(7):1655-1659.
Crossref - Weigel Linda M, Christine D Steward, Fred C Tenover. “gyrA mutations associated with fluoroquinolone resistance in eight species of Enterobacteriaceae.” Antimicrob Agents Chemother. 1998;42(10):2661-2667.
Crossref - Qiang Yan Zhi, Tong Qin, Wang Fu, et al. “Use of a rapid mismatch PCR method to detect gyrA and parC mutations in ciprofloxacin-resistant clinical isolates of Escherichia coli.” J Antimicrob Chemother. 2002;49(3):549-552.
Crossref - Park Chi Hye, Ari Robicsek, George A Jacoby, Daniel Sahm, David C Hooper. “Prevalence in the United States of aac (6′)-Ib-cr encoding a ciprofloxacin-modifying enzyme.” J Antimicrob Chemother. 2006;50(11):3953-3955.
Crossref
© The Author(s) 2020. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.