ISSN: 0973-7510
E-ISSN: 2581-690X
Acinetobacter baumannii is an opportunistic microorganism commonly found in intensive care units (ICUs), and it is responsible for a broad span of hospital-acquired infections. Persistence of nosocomial infection caused by multidrug-resistant (MDR) A. baumannii is an alarming health care issue in Egypt, and at present, colistin remains the treatment of choice for the management of MDR A. baumannii infections. A. baumannii possesses great capacity to develop and acquire resistance to a broad range of antibiotics. The acquisition and dissemination of antibiotic-resistant determinants in A. baumannii strains are mediated by integrons, especially class I integrons. This study focuses on the characterization of some genetic mechanisms underlying the multidrug-resistant phenotypes of A. baumannii isolates in Egypt. Forty-eight A. baumannii specimens were isolated from different hospitalized patients; least resistance was observed against amikacin and tigecycline, with 60% and 58.5% of the isolates resistant, respectively, whereas 62.5% of the isolates were resistant to imipenem and meropenem. The highest sensitivity was found for colistin. Genetic analysis revealed that blaoxa-51 was detected in all isolates, the blaoxa-23-like gene was detected in 80% of the isolates, and blaoxa-24 and blaoxs-58 were not detected in any isolate. Finally, PCR analysis revealed that 6.6% of isolates carried the class I integron gene.
Acinetobacter baumannii, Multidrug-resistant (MDR)
Acinetobacter baumannii is an opportunistic microorganism commonly found in intensive care units (ICUs). It is responsible for a broad range of infections such as urinary tract infections, surgical wound infections, ventilator-associated pneumonia, meningitis, bacteremia, and other life-threatening infections. The primary factor leading to the wide spread of this nosocomial microorganism is its capacity for possessing a broad spectrum of antibiotic resistance genes. Selective pressure due to environmental stress causes the prevalence of these antibiotic-resistant clones1. Its broad assortment of antimicrobial resistance genes makes infections difficult to treat, thereby causing financial burdens that ultimately create problems in infection management in hospitals. The quickly increasing prevalence of multi-drug resistant A. baumannii isolates is becoming a significant concern in global public health. The spread of resistant genes in hospitals and in society is caused by horizontal gene transfer2 and movable segments such as plasmids, transposons, and integrons are the primary genetic elements responsible for the spread and prevalence of these resistance genes3. Over the past decades, despite the discovery of new drugs and treatment choices, A. baumannii strains have been shown to possess a capability to rapidly evolve multidrug resistance (MDR). This rapid increase in MDR is not only caused by the inherent resistance genes carried by these strains, but also by their extraordinary capacity to obtain resistance elements from other microbes. The roles of efflux pumps, class B β-lactamase (metallo-β-lactamase), chromosomal class C β-lactamase, AmpC, class D β-lactamase (OXA-type carbapenemase), integrons, and associated insertion sequence elements in MDR occurrence have recently been well documented3. The acquisition and spread of antimicrobial resistance determinants in multidrug-resistant A. baumannii strains are frequently mediated by integrons, especially class I integrons. Integrons are a key element in MDR spread, particularly in gram-negative pathogens4. These are usually immobile, but they can be transported through mobile genetic elements such as plasmids and transposons. Integrons have the ability to merge into the microbial genome, and they possess numerous gene cassettes that confer antibiotic resistance5. Integrons can be categorized into three segments based on sequence conservation: the 5’-conserved segment, the variable region, and the 3’-conserved segment. The 5’-conserved segment contains a promoter, PC, and an intI gene encoding integrase; the variable region usually incorporates various gene cassettes, and the 3’-conserved segment comprises sequences acquired from transposons such as qacEΔ, sul1, and orf5, which are found in class I integrons6. Integrons carry divergent gene cassettes that are rearranged under antibiotic selective pressure7. In A. baumannii, these gene cassettes frequently comprise efflux pump genes, β-lactam resistance genes, and aminoglycoside resistance genes. Up to now, a few categories of integrons have been recognized based on the succession of the integrase gene. Among these categories, class I integrons are the most common class type, and these are fundamental in the development and spread of resistance genes, followed by class II. Reports on the different classes of integrons are scarce4,8, though based on literature, class 1 integrons and the pool of associated gene cassettes are the significant causes of A. baumannii MDR and could thus become a valuable instrument for studying molecular epidemiology in possible cross-infection cases, particularly in critical wards of hospitals such as ICUs2.
Bacterial isolates
Our study was conducted from 2016 to 2019. Forty-eight non-repeated A. baumannii isolates were collected from various clinical samples of hospitalized patients in El-Fayoum General Hospital. Isolates were identified using Gram staining, followed by specific biochemical reactions to identify Acinetobacter up to the genus level9. The isolates were then confirmed using the VITEK 2 system, following the manufacturers’ instructions10. Finally, isolates were confirmed by examination of the blaoxa_51_like gene, which is intrinsic to A. baumannii11.
Antimicrobial sensitivity testing
In vitro sensitivity tests were carried out using a panel of 11 antibiotics for all isolates using the disk diffusion method, in accordance with the guidelines established by the Clinical and Laboratory Standards Institute (CLSI)12. The following antibiotic disks were used: amoxicillin/clavulanate (AMC), piperacillin/tazobactam (TPZ), cefoxitin (FOX), cefotaxime (CTX), cefepime (FEP), imipenem (IPM), aztreonam (ATM), ciprofloxacin (CIP), tigecycline (TGC), amikacin (AK), and colistin (COL). The standard reference strain Escherichia coli ATCC 25922 was used as a quality control strain in every test run.
Determination of the minimum inhibitory concentration (MIC)
MIC was tested for each isolate using the broth dilution method, as described by Irith Wiegand13. Bacteria were cultured in Muller-Hinton broth with a serial dilution of antibiotics. Growth (turbidity) was then determined after incubation for a specific duration (16-20 h), and the MIC value was then determined.
Extraction of genomic DNA
A. baumannii isolates were refreshed by culturing in Luria-Bertani (LB) broth and incubating overnight at 37°C. Fresh culture was then used for DNA extraction by boiling 1 mL of each isolate in LB suspension in an Eppendorf tube at 100°C for 5 min in a water bath to lyse the bacterial cell wall and liberate the DNA into the medium. The tubes were then centrifuged at maximum speed for 5 min, and the supernatant was then utilized for PCR14.
Detection of integron genes in A. baumannii isolates
For mining integrons, the following primers were used: intI1, intI2, and intI3 (Table 1)15. PCR reactions were performed using 5 μL of DNA, 0.2 mM of each type of deoxynucleoside triphosphate (dNTP), and 2 μL of 10× PCR buffer. One unit of Taq polymerase, 1.5 mM MgCl2, and 1.25 μM of each primer were then added to the reaction. PCR conditions used were as follows: 35 cycles of 30 s of denaturation at 94°C, 30 s of annealing at 55°C, 30 s of extension at 72°C, with a final extension at 72°C for 10 min16.
Table (1):
PCR primers for the detection of genes encoding OXA carbapenemase and class-I, class-II and class-III integron.
| Primer name | Primer sequence | Product size | Reference |
|---|---|---|---|
| blaoxa-23like-F | GAT CGG ATT GGA GAA CCA GA | 501 bp | 17 |
| blaoxa-23like-R | ATT TCT GAC CGC ATT TCC AT | ||
| blaoxa-24like-F | GGT TAG TTG GCC CCC TTA AA | 246 bp | |
| blaoxa-24like-R | AGT TGA GCG AAA AGG GGA TT | ||
| blaoxa-51like-F | TAA TGC TTT GAT CGG CCT TG | 353 bp | 32 |
| blaoxa-51like-R | TGG ATT GCA CTT CAT CTT GG | ||
| blaoxa-58like-F | AAG TAT TGG GGC TTG TGC TG | 599 bp | 17 |
| blaoxa-58like-R | CCC CTC TGC GCT CTA CAT AC | ||
| Int1F | CAG TGG ACA TAA GCC TGT TC | 160 bp | 16 |
| Int1R | CCC GAG GCA TAG ACT GTA | ||
| Int2F | GTA GCA AAC GAG TGA CGA AAT G | 788 bp | 33 |
| Int2R | CAC GGA TAT GCG ACA AAA AGG T | ||
| Int3.F | GCC TCC GGC AGC GAC TTT CAG | 979 bp | |
| Int3.R | ACG GAT CTG CCA AAC CTG ACT |
Detection of carbapenem resistance genes
PCR was carried out in a reaction mixture containing 1× PCR buffer with 1.5 mM MgCl2, 0.2 mM dNTP, 1.5 U Taq polymerase, 3 μL DNA, and 50 pmol of forward and reverse primers per reaction.
Primer sequences used to amplify genes encoding blaoxa-23 like, blaoxa-24 like, blaoxa-51, and blaoxa-58 like genes are summarized in Table 1. The PCR conditions used to amplify OXA-type carbapenemases were as follows: initial denaturation at 94°C for 5 min, followed by 30 cycles of 94°C for 25 s, 52°C for 40 s, and 72°C for 50 s, with a final elongation step at 72 °C for 6 min17.
Bacterial isolates
All isolates showed gram-negative coccobacilli arranged in diploids. The blaoxa-51like gene was found in each isolate as a PCR product of 353 base pairs (bp).
Antibiotic sensitivity of bacterial isolates
All 48 isolates were examined. Isolates were obtained from patients with different types of infections: 29 (60%) had respiratory tract infection, 7 (15%) had blood and wound infections, 7 (15%) had urinary tract infection, and 5 (10%) had miscellaneous infections.
Antimicrobial sensitivity of A. baumannii isolates using the disk diffusion method with E. coli ATCC 25922 as the control strain showed resistance patterns as follows: (AMC): 89.6%, (TPZ): 81%, (FOX): 95.8%, (CTX): 97.9%, (FEP): 87.5%, (IPM): 62.5%, (ATM): 62.5%, (CIP): 64.5%, (TGC): 58.5%, (AK): 60.4%, and (COL): 100%.
The MIC values for carbapenems were above 32 μg/mL for (30) 62.5% of isolates, and above 128 μg/mL, 256 μg/mL and 128 μg/mL for (44) 91.6% of second-generation, (47) 97.9% of third generation, and (42) 87.5% of fourth-generation cephalosporin, respectively. Amoxicillin/clavulanate and piperacillin/tazobactam showed MIC values above 256 μg/mL for (36) 75% and (34) 70.8% of isolates, respectively. On the other hand, monobactam had values above 256 μg/mL for (31) 64.5% of isolates and above 256 μg/mL for amikacin and ciprofloxacin. Finally, colistin gave values ≤ 1 μg/mL for all 48 isolates. MIC values for the tested antibiotics are summarized in Table 2.
Table (2):
Minimum inhibitory concentration (MIC) distributions of antimicrobial agents for 48 isolates of A. baumannii.
≤0.25 |
0.5 |
1 |
2 |
4 |
8 |
16 |
32 |
64 |
128 |
≥256 |
|
|---|---|---|---|---|---|---|---|---|---|---|---|
AMC |
0 |
12 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
36 |
TPZ |
0 |
0 |
0 |
0 |
14 |
0 |
0 |
0 |
0 |
0 |
34 |
CN |
10 |
5 |
5 |
0 |
0 |
0 |
0 |
0 |
20 |
8 |
0 |
FOX |
0 |
0 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
44 |
2 |
CTX |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
47 |
CAZ |
5 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
3 |
35 |
5 |
FEP |
0 |
0 |
5 |
0 |
0 |
0 |
0 |
0 |
1 |
42 |
0 |
IPM |
0 |
0 |
0 |
12 |
1 |
3 |
2 |
30 |
0 |
0 |
0 |
MEM |
0 |
0 |
0 |
10 |
3 |
3 |
2 |
30 |
0 |
0 |
0 |
ATM |
0 |
0 |
0 |
0 |
0 |
0 |
5 |
5 |
4 |
3 |
31 |
CIP |
0 |
7 |
10 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
31 |
TGC |
0 |
7 |
8 |
0 |
0 |
0 |
0 |
4 |
20 |
9 |
0 |
AK |
0 |
8 |
10 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
30 |
COL |
3 |
5 |
40 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
AMA: Antimicrobial agent, SD: Antimicrobial serial dilution, AMC: Amoxicillin/Clavulanic acid, TPZ: piperacillin/tazobactam, FOX: Cefoxitin, CTX: ceftriaxone, CAZ: Ceftazidime, FEP: Cefepime, IPM: Imipenem, ATM: Aztreonam, CIP: Ciprofloxacin, TGC: Tigecycline, AK: Amikacin and COL: Colistin.
Integron detection in A. baumannii isolates
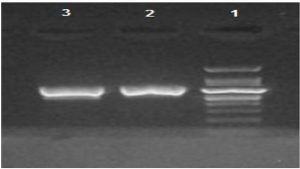
Two isolates were found to possess the class I integron gene, as shown in Fig. 1. None of the class II or class III integrons were detected.
Fig. 1. Gel electrophoresis of PCR amplified class-I integron gene in A. baumannii isolates. Lane 1 was1kb DNA ladder, lane 2 and 3 were (160 bp) amplicon of class-I integron gene positive A. baumannii isolates.
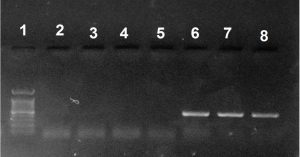
Fig. 2. Gel electrophoresis of PCR amplified blaoxa-23like gene in A. baumannii isolates. Lane 1 was 1kb DNA ladder, lane 6, 7 and 8 were (501 bp) amplicons of blaoxa-23like gene positive A. baumannii isolates while lanes 2, 3, 4 and 5 were blaoxa-23like amplicon gene negative A. baumannii.
Detection of carbapenem resistance genes
All carbapenem-resistant A. baumannii isolates were subjected to PCR screening for the presence of CHDLs. The blaoxa-51-like gene was detected in all isolates, blaoxa-23-like gene was detected in 24 isolates, and the blaoxa-24-like and blaoxa-58-like genes were not detected (Fig 2).
A total of 48 Acinetobacter clinical samples were tested for antimicrobial susceptibility, and all were found to be greater than 58% resistant to all the selected antibiotics. Ninety percent of the samples were resistant to amoxicillin-clavulanate, 96% resistant to second-generation cephalosporin, 98% resistant to third-generation cephalosporin, 88% resistant to fourth-generation cephalosporin, 62.5% resistant to carbapenems and monobactam, 64.5% resistant to quinolone, and 60.4% to resistant amikacin. In contrast, all isolates showed 0% resistance to colistin. These data indicate the extreme drug-resistant risk of nosocomial infections, consistent with data from the Middle East, published by Al-Sweih NA18 and with previous results obtained from Egypt19.
In our study, the respiratory tract was the primary site of infection, which is similar to data published by Joly-Guillou ML20. Moreover, we found that colistin is the only treatment option left for managing A. baumannii infections.
MIC values for carbapenems were above 32 μg/mL for 62.5% of isolates, and above 128 μg/mL, 256 μg/mL and 128 μg/mL for 91.6% of second-generation, 97.9% for third-generation, and 87.5% for fourth-generation cephalosporins. Amoxicillin/clavulanate and piperacillin/tazobactam showed MIC values above 256 μg/mL for 75% and 70.8% of isolates, respectively. On the other hand, monobactam revealed values above 256 μg/mL for 64.5% of isolates and above 256 μg/mL for amikacin and ciprofloxacin; these data are consistent with data from Egypt published by Fouad et al.21
Based on the obtained sensitivity and MIC data, all carbapenem-resistant A. baumannii (CRAB) isolates were subjected to further investigation.
The most common carbapenemases in A. baumannii are the carbapenem hydrolyzing class D β-lactamases (CHDLs) and metallo-β-lactamase (MBL), while Class A carbapenemases were found at a lesser extent22. CHDLs can be classified into four main subgroups: the intrinsic blaoxa-51-like and the acquired carbapenemase genes blaoxa-23-like, blaoxa-24/40like, and blaoxa-58-like17.
The blaoxa-51like gene is intrinsic to A. baumannii17,23, and its discovery in all isolates of the present study confirmed their identity, as Feizabadi, et al. observed the blaoxa-51-like gene in all 108 A. baumannii isolates in contrast to 20 negative non-A. baumannii isolates24. Moreover, Sofy et al. used the blaoxa-51-like gene for confirmation of A. baumannii isolates 25. These studies show that PCR analysis of the blaoxa-51-like can be used as a basic and reliable technique for A. baumannii recognition.
In this study, multiplex PCR showed that 24/30 (80%) of CRAB isolates harbored blaoxa-23-like genes, whereas blaoxa-24/40-like and blaoxa-58-like genes were not detected, consistent with numerous studies that reported blaoxa-23-like as the most prevalent carbapenemase among carbapenem-resistant A. baumannii 26-28. In Egypt, Fouad et al. confirmed the importance of the blaoxa23-like gene among A. baumannii isolates 21.
The class I integron has been documented in several studies of multi-drug resistant A. baumannii worldwide29. In this study, 6.6% of CRAB isolates were positive for class I integrons, while class II and class III integrons were not detected; this is consistent with a study performed in Turkey, in which 6.4% CRAB isolates were positive for class I integrons30. On the other hand, our results are not consistent with that of Mehdi et al., who reported a significant prevalence of class I and class II integrons31.
Acinetobacter baumannii poses a serious threat in Egyptian hospitals, and our current findings suggest a critical healthcare problem that can lead to the loss of effectivity of future therapeutic options. As such, stricter infection control measures are required to decrease intrahospital and interhospital spread of multi-drug resistant A. baumannii.
ACKNOWLEDGMENTS
We would like to express our heartfelt thanks to Dr Mahmoud Khalil, Department of Microbiology and Immunology, Faculty of Pharmacy Fayoum University, Fayoum, Egypt.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
ETHICS STATEMENT
Not applicable.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
- Rolain JM, Loucif L, Al-Maslamani M, et al. Emergence of multidrug-resistant Acinetobacter baumannii producing OXA-23 Carbapenemase in Qatar. New Microbes and New Infections. 2016;11:47-51.
Crossref - Ardoino I, Zangirolami F, Iemmi D, et al. Risk factors and epidemiology of Acinetobacter baumannii infections in a university hospital in Northern Italy: A case-control study. American Journal of Infection Control. 2016;44(12):1600-1605.
Crossref - Deng Y, Bao X, Ji L, Ji L, et al. Resistance integrons: class 1, 2 and 3 integrons. Ann Clin Microbiol Antimicrob. 2015;14(1):45.
Crossref - Martins N, Picao RC, Adams-Sapper S, Riley LW, Moreira BM. Association of class 1 and 2 integrons with multidrug-resistant Acinetobacter baumannii international clones and Acinetobacter nosocomialis isolates. Antimicrob Agents Chemother. 2015;59(1):698-701.
Crossref - Fluit AC, Schmitz FJ. Resistance integrons and super-integrons. Clin Microbiol Infect. 2004;10(4):272-288.
Crossref - Norrby SR, Nord CE, Finch R. Lack of development of new antimicrobial drugs: a potential serious threat to public health. The Lancet Infectious Diseases. 2005;5(2):115-119.
Crossref - Halaji M, Rezaei A, Zalipoor M, Faghri J. Investigation of Class I, II, and III Integrons Among Acinetobacter Baumannii Isolates from Hospitalized Patients in Isfahan, Iran. Oman Med J. 2018;33(1):37-42.
Crossref - Gillings MR, Gaze WH, Pruden A, Smalla K, Tiedje JM, Zhu Y-G. Using the class 1 integron-integrase gene as a proxy for anthropogenic pollution. The ISME Journal. 2015;9(6):1269-1279.
Crossref - Howard A, O’Donoghue M, Feeney A, Sleator RD. Acinetobacter baumannii. Virulence. 2012;3(3):243-250.
Crossref - Joyanes P, del Carmen Conejo Ma, Martinez-Martinez L, Perea EJ. Evaluation of the VITEK 2 System for the Identification and Susceptibility Testing of Three Species of Nonfermenting Gram-Negative Rods Frequently Isolated from Clinical Samples. J Clin Microbiol. 2001;39(9):3247-3253.
Crossref - Turton JF, Woodford N, Glover J, Yarde S, Kaufmann ME, Pitt TL. Identification of Acinetobacter baumannii by detection of the blaOXA-51-like carbapenemase gene intrinsic to this species. J Clin Microbiol. 2006;44(8):2974-2976.
Crossref - Wayne P. Clinical and laboratory standards institute. Performance standards for antimicrobial susceptibility testing. 2011.
- Wiegand I, Hilpert K, Hancock REW. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nature Protocols. 2008;3(2):163-175.
Crossref - Qi C, Pilla V, Jessica HY, Reed K. Changing prevalence of Escherichia coli with CTX-M-type extended-spectrum β-lactamases in outpatient urinary E. coli between 2003 and 2008. Diagn Microbiol Infect Dis. 2010;67(1):87-91.
Crossref - Dillon B, Thomas L, Mohmand G, Zelynski A, Iredell J. Multiplex PCR for screening of integrons in bacterial lysates. J Microbiol Methods. 2005;62(2):221-232.
Crossref - Koeleman JGM, Stoof J, Van Der Bijl MW, Vandenbroucke-Grauls CMJE, Savelkoul PHM. Identification of epidemic strains of Acinetobacter baumannii by integrase gene PCR. J Clinical Microbiol. 2001;39(1):13.
Crossref - Woodford N, Ellington MJ, Coelho JM, et al. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int J Antimicrob Agents. 2006;27(4):351-353.
Crossref - Al-Sweih NA, Al-Hubail M, Rotimi VO. Three distinct clones of carbapenem-resistant Acinetobacter baumannii with high diversity of carbapenemases isolated from patients in two hospitals in Kuwait. Journal of Infection and Public Health. 2012;5(1):102-108.
Crossref - Eman A. El-Masry HAE-M. Characterization of Carbapenem-resistant Acinetobacter baumannii Isolated from Intensive Care Unit, Egypt. Egypt J Med Microbiol. 2018;27:85-91.
- Joly-Guillou ML. Clinical impact and pathogenicity of Acinetobacter. Clin Microbiol Infect. 2005;11(11):868-873.
Crossref - Fouad M, Attia AS, Tawakkol WM, Hashem AM. Emergence of carbapenem-resistant Acinetobacter baumannii harboring the OXA-23 carbapenemase in intensive care units of Egyptian hospitals. Int J Infect Dis. 2013;17(12):e1252-e1254.
Crossref - Poirel L, Nordmann P. Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin Microbiol Infect. 2006;12(9):826-836.
Crossref - Turton JF, Woodford N, Glover J, Yarde S, Kaufmann ME, Pitt TL. Identification of Acinetobacter baumannii by detection of the blaOXA-51-like carbapenemase gene intrinsic to this species. J Clin Microbiol. 2006;44(8):2974-2976.
Crossref - Feizabadi MM, Fathollahzadeh B, Taherikalani M, et al. Antimicrobial susceptibility patterns and distribution of blaOXA genes among Acinetobacter spp. Isolated from patients at Tehran hospitals. Jpn J Infect Dis. 2008;61(4):274-278.
- Sofy KA, Saafan AE, AbdelGhani SM, Amin MA. Phenotypic and Genotypic Characterization of Different Classes of Β-Lactamases amongst Acinetobacter Spp. Isolated from Egyptian Hospitals. N Egypt J Microbiol. 2015;42.
- Al-Agamy MH, Shibl AM, Ali MS, Khubnani H, Radwan HH, Livermore DM. Distribution of β-lactamases in carbapenem-non-susceptible Acinetobacter baumannii in Riyadh, Saudi Arabia. J Glob Antimicrob Resist. 2014;2(1):17-21.
Crossref - Cicek AC, Saral A, Iraz M, et al. OXA- and GES-type β-lactamases predominate in extensively drug-resistant Acinetobacter baumannii isolates from a Turkish University Hospital. Clin Microbiol Infect. 2014;20(5):410-415.
Crossref - Al-Hassan L, El Mehallawy H, Amyes SGB. Diversity in Acinetobacter baumannii isolates from paediatric cancer patients in Egypt. Clin Microbiol Infection. 2013;19(11):1082-1088.
Crossref - Koczura R, Przyszlakowska B, Mokracka J, Kaznowski A. Class 1 Integrons and Antibiotic Resistance of Clinical Acinetobacter calcoaceticus-baumannii Complex in Poznan, Poland. Curr Microbiol. 2014;69(3):258-262.
Crossref - CiCek AC, Duzgun AO, Saral A, et al. Detection of class 1 integron in Acinetobacter baumannii isolates collected from nine hospitals in Turkey. Asian Pac J Trop Biomed. 2013;3(9):743-747.
Crossref - Goudarzi M, Azimi H. Dissemination of Classes 1, 2, and 3 Integrons in Acinetobacter baumannii Strains Recovered from Intensive Care Units Using Polymerase Chain Reaction-Restriction Fragment Length Polymorphism. Jundishapur J Microbiol. 2017;10(5):e13100.
Crossref - Brown S, Young H, Amyes S. Characterisation of OXA-51, a novel class D carbapenemase found in genetically unrelated clinical strains of Acinetobacter baumannii from Argentina. Clin Microbiol infect. 2005;11(1):15-23.
Crossref - Mazel D, Dychinco B, Webb VA, Davies J. Antibiotic resistance in the ECOR collection: integrons and identification of a novel aad gene. Antimicrob Agents Chemother. 2000;44(6):1568-1574.
Crossref
© The Author(s) 2020. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.