ISSN: 0973-7510
E-ISSN: 2581-690X
Escherichia coli are commonly found in the gastrointestinal tract of humans and other warm-blooded organisms. Since E. coli can be discharge through deposition of fecal material, it has become habituated to soil and water in the environment. Hence the present study was undertaken to isolate and identify E. coli from various environments (viz., cattle waste, piggery, poultry and sewage water) and to determine their antibiogram patterns to different group of antimicrobial agents. A total of 120 samples comprising cattle wastes (30), poultry droppings (30), piggery (30), sewage water/sediment (30) were collected from the Dakshina Kannada and analyzed for the presence of E. coli. Of total samples screened, 82 (68.3%) were found to be positive to E. coli and among the 82 E. coli strains, 25 were isolated from cattle wastes, 18 from piggery, 13 from poultry and 26 from sewage samples. The antibiogram pattern of these strains showed varied multi-drug resistance profile to the selected antibiotics. Of 82 strains, 19 (23.2%) were susceptible to all the antibiotics and 63 were resistant to at least one of the drug tested. The results from present study revealed the higher relative resistance pattern to the tested antibiotic among E. coli suggesting their potentiality in transferring MDR thereby posing public health concerns in treating problems.
Escherichia coli, antibiotic, antibiotic resistance, integrons, multi drug resistant
Antibiotics are widely used in treating infections of bacterial origin. They were predominant in treating diseases in the mid of 20th and early 21st century. Extensive use of antibiotics as medications has exerted selective pressure on vulnerable bacteria, favoring the survival of resistant bacteria1-3. These bacteria tend to take up resistance to varied antibiotics there by becomes multiple drug resistant (MDR). Infections caused by MDR bacteria are at raise posing a real threat in treating these populations. Incidence of antimicrobial resistance (AMR) is a major concern in terms of public health issue globally4. However abrupt usage of them lead to the emergence and spread of antibiotic resistance in the pathogenic organisms. The rapid emergence of antibiotic resistant bacterial strains and lack of new antibiotics to treat diseases resulted in increase in death specially in intensive care units. Thus, AMR has become a universal health threat and is a global issue. Among the food production sector, the poultry birds contribute to about 90 billion tons of meat production globally5. Due to the rapid manifestation of diseases in poultry and livestock animal there arose the question of food security. Hence usage of antibiotics has become popular in treating infections in poultry birds as well as live stocks6. Their usage in terms of growth supplements in livestock specially in farmed animals has been one of the contributing source of AMR, which are evident in various reports on isolation of MDR bacteria from livestock populations across the globe7. The global food trade is one of the leading cause of spread and colonization of resistant bacteria globally. The massive use of antibiotics in treating diseases in humans and animals has resulted in the accumulation of antibiotic compounds in the environment leading to the emergence and uptake of antibiotic resistance6. Entry of antibiotic compounds into the environment – waterbodies could be traced out from effluent discharges from hospitals and community wastes which in turn becomes the reservoir hub harboring pathogeneous resistant microorganisms7.
Certain bacterial community are inherently resistant to certain antibiotics where certain bacteria can acquire resistance by repeated exposing to group of antibiotics by mutations and by horizontal gene transfer. Various genes that are responsible for intrinsic resistance to various classes antibiotics like b-lactams, fluoroquinolones and aminoglycosides. Better understanding on the genetic basis of the bacterial intrinsic resistance along with the range of the activity of antibiotics will therefore help in choosing the combination of antibiotics to treat the bacterial infections8. The present study was aimed in isolating E. coli from different sources to analyze their genetic elements for antibiotic resistance to understand their antibiotic resistance profile, spectrum of resistance. These studies will give a preliminary idea of the transmission of AMR in E. coli.
Study design
In this study, two taluks of Dakshina Kannada district i.e., Belthangady and Mangalore of Karnataka were selected for regular sample collection. The samples (sewage water/sediment, poultry droppings, piggery and cattle wastes) were aseptically collected and brought to the laboratory for analysis. A total of 120 samples were collected from different sources – comprising 30 each from cattle wastes, poultry droppings, piggery and sewage water/sediment.
Isolation and identification of E. coli
The samples were subjected for isolation of E. coli by culture based conventional method as per the protocol recommended by FDA Bacteriological Analytical Manual9. Samples were pre-enriched in lactose broth and streaked on eosin methylene blue (EMB) agar plate. Typical colonies from EMB plate were purified and sub cultured on Luria Bertani agar (Hi Media Laboratories Pvt. Ltd., India) and subjected to a battery of biochemical tests. Identified E. coli isolates were further confirmed by molecular single step polymerase chain reaction (PCR) using genus specific primer for uidA gene (Table 1).
Antimicrobial susceptibility test
Antibiotic sensitivity profile was performed for all the identified E. coli isolates on Muller & Hinton Agar (Hi-Media Pvt. Ltd. Mumbai) using standard Kirby-Bauer disk diffusion method (Bauer et al. 1966). Antibiotic discs tested in the study were nalidixic acid (30mcg), tetracycline (30mcg), co-trimoxazole (25mcg), ciprofloxacin (5mcg), chloramphenicol (30mcg), ampicillin (10mcg), gentamicin (10mcg), nitrofurantoin (300mcg), imipenem (10mcg), meropenem (10mcg), cefotaxime (30mcg) and piperacillin-tazobactum (100/10mcg) and the values obtained were interpreted as being resistant, intermediate and sensitive according to the Clinical and Laboratory Standards Institute guidelines. A standard E. coli, ATCC 25922 (American Type Culture Collection) was used as positive control.
Detection of antibiotic resistance genes and integrons by PCR
E. coli isolate resistant to specific antibiotic was selected and tested for presence of genetic determinants responsible for resistance to particular antibiotics. Genomic DNA was extracted from the E. coli culture by CTAB (Cetyl-trimethyl ammonium bromide) method10 and used for detection of integrons and antibiotic resistance genes. PCR was carried out in 30 µl reaction mixture containing10X buffer (100mM of Tris- HCl, pH 8.3, 20mM 0f MgCl2, 500mM of KCL and 0.1% gelatin) 200mM of deoxyribonucleotide triphosphate (dATP, dTTP, dGTP and dCTP), 10 picomoles of each primer and 1 U of Taq polymerase (Bangalore Genei, Bangalore), with 2 µl of template DNA. Amplification was carried out in a MJ-Research Thermo Cycler (PTC-200, Bio-Rad) with optimized PCR programme consisted of an initial denaturation at 94°C for 5 min followed by 30 cycles with each cycle consisting of 94°C for 30 sec, Tm (annealing temperature) for 30 sec and extension for 72° C for 30 sec. The final extension was performed at 72°C for 10 min and the amplified products were resolved by 1.5 % (w/v) agarose gel electrophoresis. Different primer pairs were used for the amplification of various antibiotic resistance genes in isolates resistant to tetracycline [tetA, tetB, tetC, tetD, tetE, tetG, tetL, tetM and tetS] 11, chloramphenicol [cat1, cat2, cat3, cmlA, cmlB and floR] 11, sulphanamides [sul1, sul2 and sul3] 11, quinolones [qnrA, qnrB and qnrS] 12, ampicillin [blaTEM] and cefotaxime [blaCTXM] 11.
Multi-drug resistant E. coli strains isolated in the study were selected and examined for the presence of class 1 integron in their plasmid DNA. Plasmid was extracted by following the alkaline lysis method described by Sambrook et al. 13 and class1 integron was amplified using specific primers (Table 1) by single step polymerase chain reaction (PCR).
Isolation and identification of bacterial strains
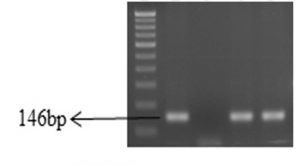
A total of 82 (68.3%) biochemically identified E. coli strains were isolated from 120 different samples (Table 2) and confirmed by amplifying 146 bp product of uidA gene with single step polymerase chain reaction (Figure 1). The incidence of E. coli from sewage was highest 86.6% followed by cattle waste-83.3%, piggery-60.0% and poultry-43.3%.
Fig. 1. Gel-electrophoresis of PCR amplified products of E. coli isolates.
Lane1: 100bp DNA ladder; Lane 2: uidA gene positive control; Lane 3: uidA gene negative control; Lane 4 and 5: Positive isolates.
Antibiotic resistance pattern
All E. coli isolates were tested for their antimicrobial susceptibility against 12 antibiotics belonging to six different groups. Of 82 isolates, 19 (23.17%) were susceptible to all the antibiotics and 63 (76.83%) showed resistance to at least one of the antibiotic tested. The incidence of antimicrobial resistant E. coli was reported highest in samples from sewage -100% (26/26) followed by poultry – 84.6% (11/13), piggery – 66.6% (12/18) and cattle wastes – 56% (14/25). The highest level of resistance was to ampicillin (56.1%), followed by nitrofurantoin (43.9%), tetracycline (41.4%), cefotaxime (37.8%), nalidixic acid (29.3%), ciprofloxacin (23.1%), co-trimoxazole (21.9%), meropenem (14.6%), imipenem (12.2), chloramphenicol (7.3%), gentamicin (4.9%), piperacillin-tazobactum (3.7%).
Detection of antibiotic resistance genes and integrons
The isolates marked resistant to tested antimicrobials were selected and analyzed for occurrence of resistant determinants of specific drug. Of 34 isolates resistant to tetracycline, seven isolates harbored more than one resistance gene (1- tetA & tetD, 1- tetA, tetC & tetS, 2- tetC & tetE, 2- tetC & tetS and 1- tetC, tetD & tetE), 4 carried tetA gene and 1 carried tetB gene. Though twenty two isolates were phenotypically resistant to tetracycline did not carried any of the genes tested. None of the isolates were harbored for tetD, tetL and tetM genes. Of six chloramphenicol resistant isolates, two carried cat1, two had cmlB and one had cmlA gene. None tested positive for cat2, cat3, and floR genes. Of 18 isolates resistant to co-trimoxazole, eight isolates harbored resistant gene tested (two- Sul1, one- Sul2, two- Sul3, two- Sul1 & Sul3 and one- Sul1, Sul2 & Sul3) and other 10 isolates did not harbored any of the tested genes. Of 46 ampicillin resistant isolates, only 2 possessed blaTEM gene and of 31 cefotaxime resistant isolates, none of them harbored blaCTX-M gene. Of total 35 quinolones resistant isolates (16- nalidixic acid resistant, 11- ciprofloxacin resistant and 8 both resistant), only one isolate harbored qnrB gene.
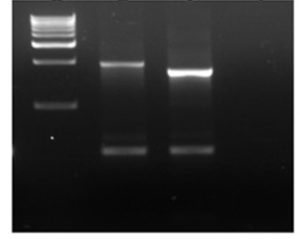
Fig. 2. Agarose gel picture of PCR amplified class1 integrons.
Lane 1: 1 kb marker, Lane 2 & 3: E. coli isolates, Lane 4: negative control.
Multi-drug resistant (MDR) isolates (resistant to more than 2 antimicrobials) were screened for the presence of integrons in their plasmid using specific primers (25). Of 24 MDR isolates, 11 isolates showed positive for class 1 integrons by amplifying products of 1610 or 1859 bp (Figure 2), in addition lower size amplicon (500 bp) was observed.
The evolution of resistance to antimicrobial agents in bacterial pathogens from different sectors is a serious threat to public health. The environment is the major harbour for antibiotic resistance, where they hold antibiotic residues which contributes for the development of resistance by selection pressure14. The irrational and inappropriate usage of antibiotics in veterinary and agriculture fields has led to the emergence and diffusion of resistance in bacteria derived from food-producing animals15,16. In particular, the emergence of antibiotic resistance in environment and microflora of food producing animals are of great concern due to the risk of getting entry into the food chain and can transmit to people and animals14,15. Hence the present study was designed to screen for antibiotic resistance E. coli from the environment (sewage) and the animal food sectors (piggery, poultry, cattle farms). Also examined for presence of genetic determinants encoding antibiotic resistance which evidences the horizontal transmission of resistance in the environment by using molecular methods like PCR. To determine the antibiogram of the isolated strains, we selected different antibiotics like ampicillin, nitrofurantoin, tetracycline, cefotaxime, nalidixic acid, ciprofloxacin, co-trimoxazole, meropenem, imipenem, chloramphenicol, gentamicin and piperacillin-tazobactum16.
In the present study, one hundred and twenty samples from different sources (sewage water/sediment, poultry droppings, piggery and cattle wastes) were screened for E. coli. Of 120 samples 82 strains were isolated, identified as E. coli by conventional methods and confirmed by molecular tools (PCR). Of 82 E. coli strains, 19 (23.17%) were susceptible to all the drug tested and 63 (76.83%) showed resistance to minimum one of the antibiotics tested in the study.
Environments are the major harbor of antibiotic-resistant microorganisms. The emergence of antibiotic resistance in microflora of food producing animals are of great concern due to the risk of getting entry into the food chain17-19. In this study, genetic determinants for antibiotic resistance were identified using molecular methods like PCR and this provided important evidences for horizontal transmission. To determine the resistance profile of the isolated strains, we selected different groups of antibiotics like ampicillin, nitrofurantoin, tetracycline, cefotaxime, nalidixic acid, ciprofloxacin, co-trimoxazole, meropenem, imipenem, chloramphenicol, gentamicin, piperacillin-tazobactum20.
Tetracyclines are the widely used therapeutic agents in human, veterinary medicines and also used in agriculture as growth promoter. In the present study, 41.4% (34/82) of isolates exhibited phenotypic resistance to tetracycline. Among the isolates from poultry practices, 8 of 13, piggery- 7 of 18, cattle wastes- 3 of 25 and sewage- 16 of 26 showed resistance to tetracycline. Earlier reports have shown that tetracycline resistant (TCr) isolates harbor genes via. tetA, B, C, D, E, G, H, L which code for efflux proteins, tetM, O, S, W which code for ribosomal protection protein (RPP), tetX which brings about enzymatic inactivation and tetU whose function is still unknown21. Of 34 tetracycline resistant isolates, seven strains harbored more than one resistance gene (1- tetA & tetD, 1- tetA, tetC & tetS, 2- tetC & tetE, 2- tetC & tetS and 1- tetC, tetD & tetE), 4 carried tetA gene and 1 carried tetB gene. Although leftover twenty two isolates were phenotypically resistant to tetracycline, none of them carried any of the resistant genes tested. This suggests that there may be a possibility of other mechanisms like enzymatic inactivation or target modification that results in resistance to tetracycline22. The tetA and tetB genes are the most common genes responsible for resistance to tetracycline and the results of this study corroborate their observation. However, the mechanisms in those isolates that did not harbour any resistance genes needs to be elucidated to be able to understand resistance transfer.
Chloramphenicol is a highly effective inhibitor of bacterial protein synthesis. Several researcher have studied the chloramphenicol resistance (3.1- 49%) in environmental isolates23,24 and is in line with the current study. In this study around 7.3% of the isolates showed resistance to chloramphenicol. Interestingly all the strains isolated from cattle wastes and sewage water system were susceptible (100%) and 1 of 18 (5.5%) isolates from piggery were resistant to chloramphenicol. Five of thirteen (38.5%) strains isolated from poultry samples were marked resistance and this may be a sign of extreme usage antibiotics (tetracycline) in poultry sector. Enzyme like chloramphenicol acetyltransferase (CAT) is responsible for resistance along with other non-enzymatic resistance gene ‘cmlA’ which codes for an efflux pump protein [25,26] and is encoded in a plasmid. Among six chloramphenicol resistant isolates, two carried cat1, two had cmlB and one had cmlA gene. None tested positive for cat2, cat3, and floR genes. The remaining one strain did not carry any of the resistance genes suggesting the possibility of other mechanism contributing to chloramphenicol resistance.
Co-trimoxazole is a compound drug which consists trimethoprim and sulfamethoxazole at 1:5 proportion and widely used to treat wide range of pathogenic bacterial infections in both humans and animals. Total 18 (21.9%) isolates showed resistant to co-trimoxazole; of which 7 of 26 from sewage, 8 of 13 from poultry, 2 of 12 from piggery and 1 of 25 from cattle wastes were phenotypically resistant. Of 18 isolates resistant to co-trimoxazole, eight isolates harbored resistant gene tested (two- Sul1, one- Sul2, two- Sul3, two- Sul1 & Sul3 and one- Sul1, Sul2 & Sul3) and other 10 isolates did not harbored any of the tested genes. Of 46 ampicillin resistant isolates, only 2 possessed blaTEM gene and of 31 cefotaxime resistant isolates, none of them harbored blaCTX-M gene. Of total 35 quinolones resistant isolates (16- nalidixic acid resistant, 11- ciprofloxacin resistant and 8 both resistant), only one isolate harbored qnrB gene. Most of the isolates (46) showed resistance to ampicillin this result were synchronized with the works of Akinbowale et al. 23 and Boujamaa24. Although 46 isolates showed resistance to ampicillin, only 2 possessed blaTEM genes, suggesting that there could be other factors contributing to resistance mechanism.
Many research findings have shown that class1 integrons were broadly distributed in many Gram negative bacterial strains from human and animals and are major site-specific recombination mechanism25,26. All the MDR isolates obtained in this study were screened for the presence of integron in that 11 isolates showed positive for class 1 integrons. Yu et al 27 reported that the occurrence of class1 integrons was strongly accompanied by resistance to various antibiotics. Further study is required to identify the genetic elements of most of the MDR isolates which is the contributing factor to uptake resistance to antimicrobials.
Acknowledgements
Financial support received from the Shri Dharmastala Manjunatheshwara Post Graduate college, Ujire toward this research work is gratefully acknowledged.
Conflict of Interest
The author(s) declare that there is no conflict of interest.
- World Organisation for Animal Health (OIE). Antimicrobials OIE, 2017. http://www.oie.int/en/ our-scientific- expertise/veterinary- products/antimicrobials.
- Food and Agriculture Organization of the United Nations. FAOSTAT: Live Animals Data. 2017. Available from: http://www.fao.org/faostat/en
- Page SW, Gautier P. Use of antimicrobial agents in livestock. Rev Sci Tech, 2012, 31:145–88.
- Maron DF, Smith TJ, Nachman KE. Restrictions on antimicrobial use in food animal production: an international regulatory and economic survey. Globalization and health, 2013, 9(1):48.
- Roca I, Akova M, Baquero F, Carlet J, Cavaleri M, Coenen S, Cohen J, Findlay D, Gyssens I, Heuer OE, Kahlmeter G, Kruse H, Laxminarayan R, Liébana E, López-Cerero L, MacGowan A, Martins M, Rodríguez-Baño J, Rolain JM, Segovia C, Sigauque B, Tacconelli E, Wellington E, Vila J. The global threat of antimicrobial resistance: science for intervention. New Microbes and New Infect, 2016, 6: 22 – 29.
- Wellington EM, Boxall AB, Cross P, Feil EJ, Gaze WH, Hawkey PM, Johnson-Rollings AS, Jones DL, Lee NM, Otten W, Thomas CM. The role of the natural environment in the emergence of antibiotic resistance in Gram-negative bacteria. Lancet Infect Dis, 2013, 13(2) :55-65.
- Jessica MA, Blair, Webber MA, Baylay AJ, Ogbolu DO, Laura J, Piddock BV. Molecular mechanisms of antibiotic resistance. Nat Rev Microbio, 2015, 13: 42-51.
- Andrews WH, Hammack T. 2011. Salmonella. In: Bacteriological Analytical Manual, Food and Drug Administration. AOAC Int , 1322-1324.
- Ausubel F, Brent RKR, Moore DD, Seidman JG, Smith JA. 1995. Current Protocols in Molecular Biology. Green Publications Associations New York, pp 121-125.
- Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular Cloning: A Laboratory Manual. Small Scale Preparation of Plasmid DNA: Alkaline Lysis Method. Cold Spring Harbor Laboratory Press, New York , pp 1.25-1.27
- Gellin G, Langlois BE, Dawson KA, Aaron DK. Antibiotic resistance of Gram-negative enteric bacteria from pigs in three herds with different histories of antibiotic exposure. Appl Environ Microbiol, 1989, 55: 2287–2292.
- Molbak K. Spread of resistant bacteria and resistance genes from animals to humans—the public health consequences. J Vet Med, 2004, 51:364–369.
- van den Bogaard AE, Stobberingh EE. Epidemiology of resistance to antibiotics. Links between animals and humans. Int J Antimicrob Agents, 2000, 14: 327–335.
- Kumar Y, Sood S, Sharma A, Mani KR. Antibiogram and characterization of resistance markers among Escherichia coli Isolates from urinary tract infections. J Infect Dev Ctries, 2013, 7(7): 513-9.
- Pinto A, Simões R, Oliveira M, Vaz-Pires P, Brandão R, da Costa PM . Multidrug resistance in wild bird populations: importance of the food chain. J. Zoo. Wildl. Med, 2015,.46(4): 723-31.
- Mittal S, Sharma M, Chaudhary U. Study of virulence factors of uropathogenic Escherichia coli and its antibiotic susceptibility pattern. Indian J Pathol Microbiol, 2014, 57(1): 61-4.
- Trevors JT, Barkay T, Bourquin AW. Gene transfer among bacteria in soil and aquatic environments: a review. Can J Microbiol, 1987, 33:191- 98.
- Chopra I, Roberts, M. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol Mol Biol Rev, 2001, 65(2): 232–260.
- French GL, Ling J, Chow KL, Mark KK. Occurrence of multiple antibiotic resistance and R-plasmids in gram-negative bacteria isolated from fecally contaminated fresh-water streams in Hong Kong. Epidemiol Infec, 1987. 98: 285-299.
- Gallardo F, Ruiz J, Marco F, Towner KJ, Vila J. Increase in incidence of resistance to ampicillin, chloramphenicol and trimethoprim in clinical isolates of Salmonella serotype Typhimurium with investigation of molecular epidemiology and mechanisms of resistance. J Medical Microbiol, 1999, 48: 367-374.
- Dorman CI, Foster TJ. Nonenzymatic chloramphenicol resistance determinants speciûed by plasmids R26 and R55-1 in Escherichia coli K-12 do not confer high-level resistance to ûuorinated analogues. Antimicrob Agent Chemother, 1982, 22:912–914.
- Cannon M, Harford S, Davies JA. A comparative study on the inhibitory actions of chloramphenicol, thiamphenicol and some ûuorinated derivatives. J Antimicrob Chemoth, 1990, 26: 307–317.
- Akinbowale OH, Peng H, Barton MD. Antimicrobial resistance in bacteria isolated from aquaculture sources in Australia. J Appl Microbiol, 2006, 100(5): 1103-1113.
- Boujamaa I. Occurrence and antibiotic resistance of mesophilic Aeromonas in three riverine freshwaters of Marrakech, Morocco. The Scientific World, 2001, 1: 796–807.
- Deekshit VK, Kumar BK, Rai P, Srikumar S, Karunasagar I, Karunasagar I. Detection of class 1 integrons in Salmonella Weltevreden and silent antibiotic resistance genes in some seafood associated nontyphoidal isolates of Salmonella in south west coast of India. J Appl Microbial, 2012. https://doi.org/10.1111/j.1365-2672.2012.05290.x
- Navajas-Benito EV, Alonso CA, Sanz S, Olarte C, Martínez-Olarte R, Hidalgo-Sanz, S, Somalo S, Torres C. Molecular characterization of antibiotic resistance in Escherichia coli strains from a dairy cattle farm and its surroundings. J Sci Food Agric., 2017. 97(1):362-365.
- Yu HS, Lee JC, Kang HY, Ro DW, Chung JY, Jeong YS, Tae SH, Choi CH, Lee EY, Seol SY, Lee YC, Cho DT. Changes in gene cassettes of class 1 integrons among Escherichia coli isolates from urine specimens collected in Korea during the last two decades. J Clin Microbio, 2003. 41:5429–33.
© The Author(s) 2018. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.