ISSN: 0973-7510
E-ISSN: 2581-690X
AmpC β-lactamases are enzymes that are resistant to β-lactams, such as penicillin and cephalosporin, but not cefoxitin and cefotetan. This study was conducted to characterize AmpC β-lactamases in Enterobacteriaceae. This study included 200 cephalosporin-resistant Gram-negative isolates recovered from different samples between January 2015 and December 2016. The isolates were subjected to phenotypic tests, and those that tested positive were further analyzed by PCR for six AmpC genotypes: ACC, DHA, FOX, CIT, MOX, and EBC. Among the 200 strains, 32% (64) were positive for AmpC β-lactamases by different phenotypic methods. The target genotypes were detected in 20 (10%) of the isolates. Pus was the predominant source of AmpC isolates. Klebsiella pneumoniae (55%) was the most common producer of AmpC β-lactamase. CIT-FOX was the predominant gene type. As there is variation in the prevalence of AmpC β-lactamases in different geographic regions, periodic surveillance and measures to control infection can prevent the spread of these genes.
AmpC β-lactamases, AmpC Disk Test, Antibiotic Resistance
Resistance to antibiotics such as cephalosporin and carbapenem, is increasingly reported in Enterobacteriaceae and is associated with the activity of several types of enzymes, notably Extended spectrum β-lactamases (ESBL), AmpC β-lactamases, and carbapenemases.
AmpC beta-lactamases are enzymes that confer resistance to penicillin, monobactam and 1st, 2nd, and 3rd generation cephalosporin as well as cephamycins.1 These enzymes are not inhibited by beta-lactam inhibitors. Sometimes also confer resistance to carbapenems along with reduced membrane permeability.2 AmpC β-lactamases are of two types: chromosomal- and plasmid-mediated. AmpC β-lactamases are chromosomally encoded in a few Gram-negative bacteria, such as Citrobacter freundii, Enterobacter spp., Proteus spp., and Serratia marcescens. Plasmid-encoded AmpC β-lactamases were first discovered in 1988.3 These were found in Klebsiella pneumonia (K. pneumoniae), Salmonella spp., and Proteus spp., through the mobility of AmpC genes in horizontal transfer between Enterobacterales.
There are no standardized guidelines for identifying AmpC β-lactamase-producing Enterobacteriaceae strains. The worldwide prevalence of plasmid-mediated AmpC ranges from 2% to 47%.4 In this study, we report the prevalence of AmpC β-lactamase-producing Enterobacteriaceae using phenotypic and molecular methods.
We carried out a prospective, lab-based cross-sectional study that included 200 randomly collected non-replicate cephalosporin-resistant Enterobacteriaceae isolates from various sample materials, including pus (n = 29), wound swab (n = 28), urine (n = 107), blood (n =11), stool (n = 2), endotracheal aspirate (ETA) secretion (n = 7), IV tip (n = 2), IV line (n = 1), and sputum (n = 13) from various departments of tertiary care (medicine, surgery, ICU, NICU, urology, nephrology, orthopedics, gynecology, pediatrics, chest and TB, special wards, and casualty) during the period from January 2015 to December 2016. Identification of the organisms were performed using standard biochemical tests.5 Antibiotic susceptibility testing was performed using the Kirby–Bauer disc diffusion method. The isolates were then subjected to phenotypic tests, namely the AmpC disc antagonism test, AmpC disc test, modified Hodge test (MHT) and phenylboronic acid test. Subsequently, phenotypic test-positive isolates were subjected to molecular characterization via multiplex PCR for the common AmpC genotypes: DHA, MOX, CIT, ACC, FOX, and EBC.
Antibiotic Susceptibility Testing6
A single colony taken from nutrient agar was inoculated into peptone water and placed in an incubator at 37°C for 2 h, and the opacity of the broth was checked with 0.5 MacFarland. Lawn cultures of the test isolates were inoculated on Muller Hinton agar (MHA) plates. Antibiotics were placed on the plates and incubated for 18-24 h at 37°C. The results were interpreted as per CLSI guidelines 2015.
Cefoxitin Screening Test6
The test isolate was taken from nutrient agar, inoculated in peptone water, and incubated for 2 h at 37°C, after which the broth turbidity was checked with 0.5 MacFarland standard. The test isolates were seeded on MHA plates. Cefoxitin discs (Himedia) were placed on surface of the MHA plates and incubated for 18-24 h. The zone of inhibition was interpreted as per CLSI guidelines 2015.
AmpC Disc Antagonism Test (DAT)7
The test isolate was inoculated in peptone water for 2 h. The opacity of the broth was checked with 0.5 MacFarland standard spread over MHA. Cefotaxime (30µg) and ceftazidime (30µg) discs were placed 20 mm apart from cefoxitin disc on MHA plates and incubated for 18-24 h. Blunting of cefotaxime or ceftazidime inhibition zone near cefoxitin was considered a positive isolate for the AmpC test.
Modified (Cefoxitin) Hodge Test (MHT)8
E. coli ATCC 25922 was seeded on MHA. Cefoxitin (30μg) discs was placed on the MHA. The isolates was inoculated from the edge of cefoxitin disc until the periphery and incubated at 37°C overnight. Isolates with cloverleaf patterns were positive for AmpC production.
AmpC Disc Test9
E.coli ATCC 25922 lawn culture was inoculated on an MHA plate. Cefoxitin disc was placed on MHA. Saline solution (20µL) was added to a sterile plain disc and placed near the cefoxitin disc. Three to four colonies of the strain were placed on the plain disc. The plate was incubated at 35°C for 18 h. The plates were read as follows: indentation (heart-shaped)/flattening of the zone around cefoxitin disc was positive for ampC production and no distortion around cefoxitin disc indicated a negative result.
Phenyl Boronic Acid Test10
Phenyl boronic acid (Himedia) (120 g) was dissolved in three mL of dimethylsulfoxide. Sterile water (3 mL) was added to this solution. Twenty microliters of the prepared phenyl boronic acid were pipetted onto cefoxitin discs. The discs were then dried for 30 min. The test strains were seeded on MHA, air-dried and cefoxitin (30 μg) and discs containing cefoxitin/phenylboronic acid (30/400 μg) were placed on the plate. The seeded plates were then incubated for 18 h at 35°C. An organism that produces an enhanced zone of inhibition of 5 mm or more around the boronic acid/cefoxitin disc compared to the zone of inhibition around the cefoxitin disc was considered an AmpC-producing organism.
Multiplex PCR for AmpC β- lactamases
Preparation of Template DNA
All 64 isolates that were positive for the ampC disc test and phenylboronic acid were subjected to multiplex PCR for amplification.11
The isolates were grown on nutrient agar and the colonies were suspended in microcentrifuge tubes containing 500 µL of sterile water. The tubes were kept in a water bath at 95°C for 10 min for cell lysis. The cells were further centrifuged for 15 min at 10000xg to remove cellular debris. The supernatant was used as a DNA template. Multiplex PCR was performed using an Eppendorf thermocycler. The primers used were designed by Perez and Perez and Hansen.11 The PCR products were analyzed using 2% agarose gel electrophoresis and visualized using a gel documentation device. A molecular ladder of 100bp was used as the marker.
Analysis
The data were analyzed using the Statistical Package for the Social Sciences (SPSS) Version 20, using simple descriptive statistics, percentiles and qualitative analysis.
Of the 200 isolates, 149 were found to be resistant to cefoxitin (i.e., results were positive for cefoxitin screening). Of these 149 isolates, 64 (42.9%) were Escherichia coli (E. coli), 65 (43.5%) K. pneumoniae, 9 (6.0%) Klebsiella oxytoca, 4 (2.6%) Proteus mirabilis (P. mirabilis), 2(1.3%) Proteus vulgaris (P. vulgaris), 1(0.7%) Citrobacter spp., and 4(2.6%) Enterobacter spp. The resistogram of the isolates indicated: 94.5% resistant to ceftriaxone, 69% to cefepime, 28% to cefepime-tazobactam, 29.5% to piperacillin-tazobactam, 32.5% to imipenem, 31.5% to meropenem, 41.5% to norfloxacin, 79% to ciprofloxacin, 41.5% to nitrofurantoin, 4% to fosfomycin, 65.5% to gentamycin, 48% to amikacin, 37% to chloramphenicol and 70% to cotrimoxazole.
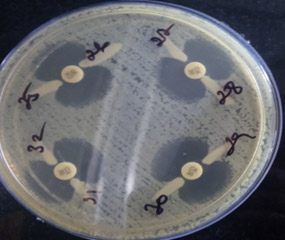
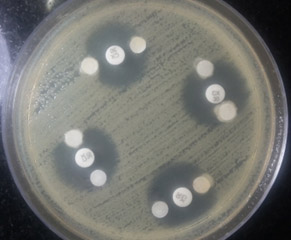
Among the 200 isolates, 62 (31%) were found to be positive for AmpC β-lactamases by the AmpC disc test (Figure 1) and MHT test (Figure 2), and 64 (32%) isolates were positive by the phenylboronic acid method. The phenotypically positive isolates are listed in Table 1.
Table (1):
Phenotypic AmpC Positive isolates Test.
No. |
Organism |
No of isolates |
AmpC producers positive in AmpC disk & MHT test |
Ampc producers by phenylboronic acid test |
|---|---|---|---|---|
1 |
Escherichia coli |
87 |
24 (27.5%) |
25 (28.7%) |
2 |
Klebsiella pneumoniae |
84 |
31(36.9%) |
32 (38.0%) |
3 |
Klebsiella oxytoca |
13 |
1 (7.6%) |
1(7.6%) |
4 |
Proteus mirabilis |
7 |
2 (28.5%) |
2 (28.5%) |
5 |
Proteus vulgaris |
3 |
2 (66.7%) |
2 (66.7%) |
6 |
Citrobacter |
1 |
1(100%) |
1 (100%) |
7 |
Enterobacter |
5 |
1 (20%) |
1(20%) |
Total |
200 |
62 (31%) |
64 (32%) |
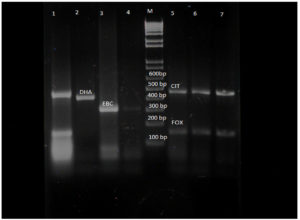
The positive isolates from the phenotypic AmpC test were subjected to multiplex PCR using primers for six families. In Table 2 we present the AmpC producers identified by multiplex PCR. Twenty isolates were assigned the genotypes DHA, FOX, CIT, and EBC (Figure 3). The majority of organisms were found to be positive for FOX-CIT. The distribution of AmpC producers in the various samples is shown in Table 3.
Table (2):
AmpC producer confirmation by multiplex PCR.
No |
Organism |
No of organism |
pAmpC producers |
|---|---|---|---|
1 |
E. coli |
87 |
8 |
2 |
K. pneumoniae |
84 |
11 |
3 |
K. oxytoca |
13 |
0 |
4 |
P. mirabilis |
7 |
1 |
5 |
P. vulgaris |
3 |
0 |
6 |
Citrobacter |
1 |
0 |
7 |
Enterobacter |
5 |
0 |
Total |
200 |
20 |
Table (3):
Distribution of plasmid mediated AmpC genes in the study isolates.
Microorganism |
pAmpc negative |
FOX-CIT |
FOX |
EBC |
DHA |
pAmpC Genotypes |
|---|---|---|---|---|---|---|
E. coli (25) |
17 |
7 |
0 |
1 |
0 |
8 (40%) |
K. pneumoniae (33) |
22 |
9 |
1 |
1 |
0 |
11 (55%) |
K. oxytocoa (1) |
1 |
0 |
0 |
0 |
0 |
0 |
P. mirabilis (2) |
2 |
0 |
0 |
0 |
1 |
1(5%) |
P. vulgaris (1) |
1 |
0 |
0 |
0 |
0 |
0 |
Enterobacter spp (1) |
1 |
0 |
0 |
0 |
0 |
0 |
Citrobacter spp (1) |
1 |
0 |
0 |
0 |
0 |
0 |
Total (64) |
44 |
16 |
1 |
2 |
1 |
20 |
Figure 3. Gel electrophoresis of AmpC beta lactamases producers. Lane 1,5,6,7 – FOX-CIT, Lane M – molecular ladder (100bp), Lane 2 – DHA, Lane 3, 4 – EBC
Pus and wound samples were the predominant sources of AmpC. Klebsiella pneumoniae is the predominant AmpC producer. (Table 4)
Table (4):
Distribution of AmpC producers among the different samples by Multiplex PCR.
No |
Sample |
Total no of samples |
E.coli |
AmpC producers |
Klebsiella pneumoniae |
AmpC producer |
Proteus mirabilis |
AmpC producer |
|---|---|---|---|---|---|---|---|---|
1 |
Urine |
107 |
51 |
4 |
41 |
5 |
3 |
0 |
2 |
Pus |
29 |
16 |
3 |
11 |
3 |
1 |
0 |
3 |
wound |
28 |
10 |
1 |
9 |
2 |
1 |
1 |
4 |
ETA |
7 |
0 |
0 |
6 |
1 |
1 |
0 |
200 |
77 |
8 |
67 |
11 |
6 |
1 |
Treatment failures due to broad-spectrum antibiotics may be due to over expression of AmpC β-lactamases. AmpC β-lactamases encoded by plasmids were detected more frequently in E. coli and Klebsiella species.12 Several studies have revealed discrepancies in the types of AmpC β-lactamases among different geographical regions. AmpC beta-lactamase prevalence in different parts of India ranges from 8% to 47%.13-17 The AmpC producer rate was 32% in our study, which is similar to the results of other studies.18-19
In our study, all isolates were negative for the disc antagonism test (DAT), thus revealing the presence of only plasmid-mediated resistance in our isolates.
Cefepime is generally sensitive to AmpC producers. However, in our study, a few AmpC isolates were resistant to cefepime and cefepime/tazobactam. This may be due to a point mutation in the active site R2 loop, which results in variation that may act on cefepime in AmpC β-lactamase isolates and is known as extended-spectrum cephalosporinases.20 Many of the isolates in the present study were multidrug-resistant (MDR) because the genes are encoded on very large plasmids that are responsible for the multiresistant phenotype.
Presently, there are no recommended methods (CLSI guidelines) for screening and identification of AmpC producers in Enterobacteriaceae; however, several detection methods have been developed, such as the AmpC disc test, modified three-dimensional test, modified hodge test and inhibitor-based methods such as cloxacillin and boronic acid tests. A reduction in susceptibility to cefoxitin antibiotic is considered a screening method for the detection of AmpC enzyme. However, ACC types are the only AmpC enzymes that can be missed when using the cefoxitin screening test.
In this study, all 200 Enterobacteriaceae isolates that were not susceptible to oxyimino-cephalosporin were screened for cefoxitin susceptibility. Of the 200 isolates, 149 (74.5 %)were resistant to cefoxitin. However, only 62 (31%) out of 200 isolates were positive in the AmpC disc test and MHT, and 64 (32%) were positive in the phenylboronic acid test. The cefoxitin-resistant isolates that were AmpC-negative in the phenotypic test may be due to ESBL or MBL production, or it may be due to decreased porin channels or increased efflux pump action, as demonstrated by porin Omp K 35 and Omp K36 loss in E. coli and K. pneumoniae isolates.21 This may also be due to the over expression of chromosomal AmpC genes caused by mutations. In several other studies conducted in India,14,22,23 19-27% of cefoxitin-resistant isolates were found to be non-producers of AmpC β-lactamases. Porin loss is found to enhance the resistance of ESBL and AmpC β-lactamases and also leads to resistance to carbapenems.
The percentage of AmpC-producing isolates in the clinical samples was higher in samples from inpatients, indicating its nosocomial importance.
Among the AmpC disc and MHT tests, 24 isolates (38.7%) were E. coli, 31 (36.9 %) were K. pneumoniae. There was one isolate each of K. oxytoca, Citrobacter spp., and Enterobacter spp. As well, two isolates each were from P. mirabilis and P. vulgaris. In other studies, 24% and 20.5% of E. coli were positive in the AmpC disc test.24,25
Among the 64 AmpC-positive isolates tested, AmpC genes were detected by PCR in 20 isolates (10%). The FOX-CIT family member was detected in 16 isolates (80%). FOX-CIT was detected in nine isolates of K. pneumoniae and seven of E. coli. The genotypes detected included DHA, EBC, CIT and FOX, respectively. MOX and ACC were not detected in the present study.
AmpC β-lactamases mediated by plasmids were detected more frequently in E. coli and Klebsiella species.12 Several studies have revealed discrepancies in the types of AmpC β-lactamases among different geographical regions. The discrepancy between phenotypic and genotypic tests may be due to the presence of other AmpC β-lactamase genes that continue to expand apart from the six gene families detected by PCR,26 or may be due to hyper producers due to over expression of the chromosomal gene.26 Of the 20 isolates identified as possessing plasmid-mediated AmpC, the sources were from urine, pus, and ETA samples.
Among the 64 isolates of AmpC producers from the phenotypic test, 20 could be assigned a genotype, which included CIT-FOX (16), EBC (2), FOX (1), and DHA (1).
AmpC beta-lactamases were identified in the urine (n=9), pus (n=6), wound (n=4), and ETA (n=1). Of these samples, 80% were from inpatients. Overall, the prevalence of AmpC genotypes among the tested Enterobacteriaceae was low (10%). FOX-CIT (80%) was the predominant genotype detected in this study. FOX-CIT was more common in K. pneumoniae (45%) than in E. coli, and EBC was detected in K. pneumoniae and E. coli, one DHA from P. mirabilis, and one FOX in K. pneumoniae.
In our study, the FOX group (n=17) was predominant (85%), followed by the CIT (n=16) (80%), EBC (n=2 (10%), and DHA (n=1(5%) groups.
In a 2012 study by Mohamudha et al.,15 the DHA gene was predominant, whereas, in the study by Manoharan et al.,18 FOX-CIT was predominant, as was the case with the current study.
In our study, 40% of E. coli and 55% of Klebsiella pneumoniae were AmpC producers among the 64 phenotypic type positive clinical isolates, which is in accordance with the results of previous studies.27,28 However, 25.5% of E. coli and 39.1% of K. pneumoniae isolates have been reported in one study.29
Plasmid-mediated AmpC β-lactamases were obtained from nine urine samples, six pus samples, four wound samples, and one ETA sample. Of these, 80% were from inpatients. The overall prevalence of common plasmid-mediated AmpC genotypes among the tested Enterobacteriaceae was low (10%). In the present study, CIT-FOX (80%) was the predominant genotype. In our study, the FOX group (n=17) was predominant (85%), followed by the CIT (n=16) (80%), EBC (n=2 (10%), and DHA (n=1(5%)) groups. In a study by Mohamudha et al.15 the DHA-CIT genotype was predominant, whereas, in Manoharan et al.,18 the FOX-CIT genotype was predominant, which is in concordance with our study.
Many studies have shown that pAmpC β-lactamase-producing organisms are detected in prolonged hospitalized patients, and very recent studies suggest that it is found in tertiary care centers and outpatients, indicating its presence in the community. In this study, pAmpc was isolated from both inpatients and outpatients, indicating its presence both in hospitals and in the community.
In this study, E. coli, K. pneumoniae and P. mirabilis showed the presence of plasmid-mediated AmpC β-lactamases by PCR. In this study, many cefoxitin-resistant isolates were negative for AmpC, indicating the presence of other resistance mechanisms. FOX-CIT was the predominant AmpC β-lactamase gene, followed by EBC. The broad use of cephalosporins in empirical therapy in Enterobacteriaceae has increased the selective pressure among AmpC lactamase-generating isolates, increasing its prevalence. Hence, the identification of these enzyme-producing isolates will help in understanding the epidemiology of resistance mechanisms.
ACKNOWLEDGMENTS
The authors would like to thank Sree Balaji Medical college and Hospital, chrompet, BIHER, Chennai, India for providing the support to complete this work.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct, and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analysed during this study are included in the manuscript.
ETHICS STATEMENT
The study was conducted according to the guidelines of Institutional Human ethical committee, SBMCH, India with reference number 002/SBMC/IHEC/2013-99
- Kameyama M, Yabata J, Nomura Y, Tominaga K. Detection of CMY-2 AmpC β-lactamase-producing enterohemorrhagic Escherichia coli O157:H7 from outbreak strains in a nursery school in Japan. J Infect Chemother. 2015; 21:544-546.
Crossref - Philippon A, Arlet G, Jacoby GA. Plasmid-determined AmpC-type β-lactamases. Antimicrob Agents Chemother. 2002; 46:1-11.
Crossref - Varghese DS, Sekar U, Shanthi M, et al. Concurrent occurrence of Amp C and Cefotaxime (CTX)-M inclinical isolates of enterobacteriaceae. J Acad Clin Microbiol. 2014;16:11-6.
Crossref - Bauernfeind A, Chong Y, Lee K. Plasmid-encoded AmpC β-lactamases: how far have we gone 10 years after the discovery? Yonsei Med J. 1998; 39:520-525.
Crossref - PR Murray, EJ Baron, JH Jorgensen, et al. Manual of Clinical Microbiology, American Society for Microbiology, Washington, DC, USA, 9th edition, 2007.
- Clinical Laboratory Standards Institute. Performance standards for Antimicrobial Susceptibility Testing: Twenty first Informational Supplement. CLSI document M100-S21. Wayne: Clinical and Laboratory Standards Institute; 2015.
- Upadhyay S, Sen MR, Bhattacharjee A. Presence of different β-lactamase classes among clinical isolates of Pseudomonas aeruginosa expressing AmpC β-lactamase enzyme.
J Infect Dev Ctries. 2010; 4(4): 239 242.
Crossref - Yong D, Park R, Yum Jh et al. Further Modification of the Hodge Test to Screen Ampc Β- Lactamase (CMY-1)-Producing Strains of Escherichia Coli and Klebsiella pneumoniae. J Microbiol Methods. 2002;3:407-10.
Crossref - Singhal S, Mathur T, Khan S et al. Evaluation of the methods for AmpC β-lactamase in gram negative clinical isolates from tertiary care hospitals. Ind J Med Microbio. 2005; 23:120-124.
Crossref - Tan TY, Ng LS, He J, Koh TH, Hsu LY. Evaluation of screening methods to detect plasmid- mediated AmpC in Escherichia coli, Klebsiella pneumoniae and Proteus mirabilis. Antimicrob Agents Chemother. 2009;53:146-9.
Crossref - Pérez-Pérez FJ, Hanson ND. Detection of plasmid mediated AmpC β-lactamase genes in clinical isolates by using multiplex PCR. Journal of Clinical Microbiology. 2002; 40: 2153-2162.
Crossref - Woodford N, Reddy S, Fagan EJ et al. Wide geographic spread of diverse acquired AmpC beta-lactamases among Escherichia coli and Klebsiella spp. in the UK and Ireland. J Antimicrob Chemother, 59(1):102-5.
Crossref - Shahid M, Sobia F, Singh A, Khan HM. Concurrent occurrence of blaampC families and blaCTX-M genogroups and association with mobile genetic elements ISEcp1, IS26, ISCR1, and sul1-type class 1 integrons in Escherichia coli and Klebsiella pneumoniae isolates originating from India. J Clin Microbiol. 2012;50:1779-82.
Crossref - Sinha P, Sharma R, Rishi S, et al.Prevalence of extended spectrum beta lactamase and AmpC beta lactamase producers among Escherichia coli isolates in a tertiary care hospital in Jaipur. Indian J Pathol Microbiol. 2008;51(3):367-9.
Crossref - Mohamudha PR, Harish BN, Parija SC. Molecular description of plasmid-mediated AmpC ß-lactamases among nosocomial isolates of Escherichia coli & Klebsiella pneumoniae from six different hospitals in India. Indian J Med Res. 2012;135(1):114-9.
Crossref - Shanthi M, Sekar U, Arunagiri K, Sekar B. Detection of Amp C genes encoding for beta-lactamases in Escherichia coli and Klebsiella pneumoniae. Indian J Med Microbiol. 2012;30:290-5.
Crossref - Upadhyay S, Sen MR, Bhattacharjee A. Presence of different β-lactamase classes among clinical isolates of Pseudomonas aeruginosa expressing AmpC β-lactamase enzyme. J Infect Dev Ctries. 2010;4:239-242.
Crossref - Manoharan A, Sugumar M, Kumar A et al. Phenotypic & molecular characterization of AmpC ß-lactamases among Escherichia coli, Klebsiella spp. & Enterobacter spp. from five Indian Medical Centers. Indian J Med Res. 2012;135(3):359-64.
- Mutasim E Ibrahim, Abbas M, Al-Shahrai A, Elamin BK. Phenotypic Characterization and Antibiotic Resistance Patterns of Extended-Spectrum ß-Lactamase- and AmpC ß-Lactamase-Producing Gram-Negative Bacteria in a Referral Hospital, Saudi Arabia. The Canadian journal of infectious diseases & medical microbiology. 2019:6054694.
Crossref - Mandell, Douglass & Bennett’s functional group. Priniciples & practice of infectious diseases. 2015; Eight Edition.
- Ananthan S and Subha A. Cefoxitin resistance mediated by loss of a porin in clinical strains of Klebsiella pneumoniae and Escherichia coli. Indian J Med Microbiol. 2005; 23(1): 20-23.
Crossref - Easwaran S, Yerat RC, Ramaswamy R. A study on detection of extended-spectrum beta-lactamases (ESBLs) and comparison of various phenotypic methods of AmpC detection in Pseudomonas aeruginosa from various clinical isolates in a tertiary care teaching hospital. Muller J Med Sci Res. 2016;7:35-9.
Crossref - Manchanda V and Singh NP. Occurrence and detection of AmpC beta-lactamases among Gram-negative clinical isolates using a modified three-dimensional test at Guru Tegh Bahadur Hospital, Delhi, India. J Antimicrob Chemother. 2003;51(2):415-8.
Crossref - Bagali SO, Peerapur BV. Detection of AmpC Beta-lactamases among Escherichia coli isolates at a tertiary care hospital in Karnataka. Al Am een J Med Sci. 2013;6(1):85-87.
- Shivanna V, Rao A. Detection of AmpC β-lacatmases among the Gram Negative Clinical isolates. International journal of Recent Trends in Science and Technology. 2014;9(3):361-364.
- Wassef M, Behiry I, Younan M, Guindy NE, Mostafa S, Abada E. Genotypic Identification of AmpC ß-Lactamases Production in Gram-Negative Bacilli Isolates. Jundishapur journal of microbiology. 2014;7(1), e8556.
Crossref - Akujobi CO, Odu NN, Okorondu SI. Detection Of Amp C Beta Lactamases In Clinical Isolates of Escherichia coli And Klebsiella. Afr. J. Cln. Exper. Microbiol. 2012;13(1): 51-55.
Crossref - Altun S, Tufan ZK, Yağcı S, et al. Extended spectrum beta-lactamases, AmpC and metallo beta-lactamases in emerging multi-drug resistant Gram-negative bacteria in intensive care unit. Sci Rep. 2013;2(4):707
- Parveen RM, Harish BN, Parija SC. AmpC Beta lactamases among gram negative clinical isolates from a tertiary hospital, South India. Braz J Microbiol. 2010;41(3):596-602.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.