ISSN: 0973-7510
E-ISSN: 2581-690X
Bronchoalveolar lavage (BAL) is the saline wash of the bronchial tree, which aids in diagnosing various pulmonary pathologies. The present study was contemplated with an aim to know the clinical, microbiological profile of BAL samples along with its sensitivity pattern and to assess its utility as a diagnostic tool. This was a prospective observational study, carried over 90 patients presenting with lower respiratory tract infections. The total microbial recovery rate from BAL was 39 (43.3%). The sensitivity, specificity and positive predictive value of BAL were found to be 76.4%, 89.7% and 90.6% respectively. Maximum isolates were bacteria (25.5%) followed by Mycobacterium tuberculosis (MTB) (16.6%) and fungi (1.1%). Predominant bacterial isolates were Gram-negative (81.5%) compared to Gram-positive (18.5%). Multidrug resistance (MDR) in bacteria was seen in 59.2% of isolates. BAL is a valuable diagnostic tool to find not only bacterial but mycobacterial and fungal infections in patients with lower respiratory tract infection (LRTI). A trend towards LRTI with Gram-negative infections is on the rise and they tend to be multidrug-resistant. Hence checking susceptibility patterns is crucial to start evidence-based treatment.
Bronchoalveolar lavage (BAL), LRTI, MTB, MDR, Bacteria, Fungi
Bronchoalveolar lavage (BAL) was introduced as a research-oriented procedure way back in 19701. It was originally used as a tool for obtaining secretions and cells from the lower respiratory tracts of patients with interstitial, occupational, or both types of pulmonary diseases2. It was used for first time in India in 1994 for its important role in diagnosis of infections and malignancies3. Lower Respiratory tract infections (LRTI), caused by various organisms like bacteria and fungi, can be isolated by culturing BAL samples4. Sputum samples are still considered effective in diagnosing infective lung conditions in our country, as they are easy to obtain. Although they have 24% diagnostic yield, contamination of sample from oral flora during expectoration makes it unsatisfactory for culture5. BAL has an advantage compared to repeat sputum sample collections as the contamination by oral flora can be avoided easily. Further, the quantitative culture with proper colony cut off provides a greater insight for therapy. Another distinct advantage of BAL is that being an outpatient procedure, it is very much useful in suspected cases of community acquired pneumonia (CAP). Several studies from western literatures show that BAL can be used to increase microbial isolation6. LRTI needs to be diagnosed with appropriate diagnostic tool for evidence-based management of patients. The clinical features of respiratory diseases vary with age, gender, season, associated risk factors and supplementary factors like environment and host7. As there are only few prospective studies reported from our country investigating the role of BAL in microbial isolation, the present study was undertaken to ascertain the clinical and microbiological profile of broncho alveolar lavage samples. We have also made an attempt to determine the susceptibility pattern, which can guide the clinicians to practice evidence-based management of LRTI.
The present study was carried out with an aim to find out the microbiota of BAL samples from patients of LRTI. The objective was to study the microbial profile of BAL samples, its utility as a diagnostic tool and to identify the divergent microbiota along with antimicrobial susceptibility pattern.
This observational prospective study was carried out in a study population of 90 patients in the Department of Microbiology, Mamata medical college, Khammam, Telangana, over a period of 01 year from April 2018 to April 2019 on both outpatients as well as admitted indoor patients in the Department of Pulmonology in Mamata General Hospital, affiliated to Mamata Medical College. The hospital mainly caters the rural population in the region. Institutional Ethical Committee (IEC) clearance was obtained before start of the study. The study was started initially as a short-term studentship (STS) project of Indian Council of Medical Research (ICMR – Ref No.2018-00184) and was further continued for total duration of 1 year. The inclusion criteria of the study were patients between 14 to 80 years of age presenting with symptoms of LRTI who were negative on sputum microscopy/culture and patients willing to participate in study. Exclusion criteria were patients below 14 years and above 80 years of age; patients with lung malignancy; unstable cardiac conditions and those who did not give consent to undergo the procedure.
The present study was planned in collaboration with Department of Pulmonology. Patients presenting with symptoms of LRTI like fever and cough, with or without expectoration, shortness of breath and chest pain ≤2 weeks were assessed with thorough history and physical examination. Relevant blood and radiological investigations in the form of chest X – ray was done in every case. As study protocol, sputum/Induced sputum samples of all the patients were sent to microbiology lab. Gram stain was performed initially followed by Bartlett’s grading to check sputum sample quality8. Zeihl-Neelson (ZN) stain was performed as per Revised National Tuberculosis Control Programme (RNTCP) guidelines. KOH mount was done in suspected cases of fungal etiology. Bacterial culture was done on blood agar, macconkey agar, chocolate agar and incubated at 37°C. Chocolate agar was incubated in candle jar for 24-48 hours to check the growth of fastidious bacteria like Haemophilus influenzae, Haemophilus parainfluenzae and Moraxella catarrhalis9. Further identification of isolates was done by routine biochemical tests. Sabourauds dextrose agar (SDA) was used for fungal culture. Patients, in whom sputum/ induced sputum samples showed non satisfactory growth / no pathogenic organism/ mixed flora, were further followed up for a period of 03 days. If symptoms persisted during follow up, BAL was performed in all those cases. These were the patients with diagnostic dilemma as clinical, radiological (chest X-ray) and microbiology report were not analogous. High resolution computerised tomography (HRCT) was also done in those patients who had inconclusive evidence on chest X-ray10.
A standard BAL procedure was done in bronchoscopy room with 5 mm flexible fibre optic bronchoscope under sterile conditions. 10% lignocaine spray was used for local anaesthetic effect. Minimal sedation was used only in anxious patients. Access to BAL was achieved through nasal approach. BAL aliquots of 20 mL were used in 0.9% saline via syringe up to a total volume of 120-150 mL. Suction pressure of 100 mm of H2O was applied and about 70% of total instillation was retrieved. This fluid was then immediately sent to microbiological laboratory for examination.
Samples were subjected to Gram’s stain for quality check. Samples were not processed further if more than 1% epithelial cells were seen on low power magnification, along with absence or presence of bacteria in less than than 1-2/ oil immersion field which was suggestive of oropharyngeal contamination11,12. On accepted samples Ziehl-Neelsen (ZN) staining was done for Mycobacterium tuberculosis (MTB). KOH mount was done for fungal elements. These were preliminary microscopy procedures on the sample.
The sample was inoculated on Blood agar, MacConkey agar and chocolate agar using a sterile 4 mm nichrome loop (0.01ml) and incubated at 37°C for 18-24 hours for quantitative bacterial culture using standard laboratory techniques. Chocolate agar was placed in candle jar for 48-72 hrs to look for fastidious organisms. Sample was also inoculated on Sabaroud’s dextrose agar (SDA) for fungal culture and incubated at 37°C for 04 weeks for growth. For MTB detection, Cartridge Based Nucleic Acid Amplification Test (CBNAAT) was performed as per RNTCP guidelines.
For bacterial growth on culture plates, the colony forming unit was calculated with cut off > 105 CFU/ ml13. Colonisers were ruled out from true pathogens by strict diagnostic criteria if colony count was significant14. Antibiotic sensitivity testing was done on bacterial isolates as per Clinical and Laboratory Standards Institute (CLSI) 2018-19 guidelines by Kirby-Bauer’s disc diffusion method along with local antibiogram guidelines as per institute protocol.
Definition
Pneumonia as in LRTI was diagnosed as patients with a chest radiograph finding suggestive of new/progressive/persistent (>24hours) infiltrate and with minimum of 02 criteria like purulent expectorant; body temperature of > 38.5°C or < 36.5°C; WBC count of >10,000 cells/µL or <4,000 cells/µL and breathlessness. HRCT evidence of a segment involvement, peribronchovascular or scattered ground glass or reticular opacity or consolidation congruent with lung involvement in acute phase was considered as evidence of pneumonia. Along with this Quantitative BAL cultures of >105 CFU/ml were considered as diagnostic of LRTI.
Multidrug-resistant bacteria (MDR)7,15
Multidrug-resistant (MDR) bacteria were S. pneumoniae resistant to penicillin and other broad-spectrum agents like fluoroquinolones and macrolides, Methicillin resistant S. Aureus (MRSA) and MDR Gram negative bacilli. MDR Gram negative bacilli were Escherichia coli, Klebsiella spp., Enterobacter spp and Citrobacter spp, which were resistant to at least 03 of the following group of antibiotics,viz; piperacillin or ampicillin/sulbactam, aminoglycosides, third or fourth generation cephalosporins, carbapenems and fluoroquinolones. Pseudomonas aeruginosa resistant to at least 03 of the following antibiotic groups was also classified as an MDR,viz; piperacillin, aminoglycosides, cephalosporins, fluoroquinolones and carbapenems. Acinetobacter baumannii resistant to most of the antimicrobial agents with/without imipenem resistance was considered as MDR.
Statistical analysis
Data was entered in Microsoft excel sheet. For analysis of continuous data, unpaired t test was used. For analysis of categorical data Pearson’s ϰ2 test was used. Differences were considered statistically significant, if p<0.05. IBM SPSS Statistics for Windows, version 24 (IBM Corp., Armonk, N.Y., USA) software program was used for statistical calculations.
Study sample consisted of 90 patients who were clinically suspected but were sputum negative/unsatisfactory. Clinical profile of patients is shown in Table 1 with 61 (67.7%) males and 29 (32.2%) females with mean age of 43 years. Predisposing factors like smoking was seen in 32 (35.5%), diabetes in 23 (25.5%), chronic bronchitis in 34 (37.7%), past history of tuberculosis in 11 (12.2%) and chronic obstructive pulmonary disease (COPD) in 29 (32.2%) patients. Dyspnoea was the most common symptom (92.2%) followed by fever (76.6%) and cough with expectoration (75.5%). In patient treatment was needed for 22 (24.4%) patients.
Table (1):
Clinical profile of patients (n=90).
| Variables | Total (%) |
|---|---|
| Male | 61 (67.7%) |
| Female | 29 (32.2%) |
| Mean Age(yrs) | 43 (14-80) |
| Smoking history | 32 (35.5%) |
| Comorbidity | |
| Diabetes | 23 (25.5%) |
| Chronic Bronchitis | 34 (37.7%) |
| COPD* | 29 (32.2%) |
| Previous TB | 11 (12.2%) |
| Symptoms/Signs | |
| Fever | 69 (76.6%) |
| Sputum | 68 (75.5%) |
| Dyspnea | 83 (92.2%) |
| Leukocytosis/Leucopenia | 41 (45.5%) |
| IPD** | 22 (24.4%) |
*Chronic Obstructive Pulmonary disease, **In patient department
Clinical diagnosis of LRTI could be achieved in 51 patients as per predefined criteria. 39 (76.4%) of those 51 patients had colony count of >105 CFU/mL, with true pathogens (Mycobacteria and fungi included) on BAL fluid culture. 0f 90 patients 39 (43.4%) patients did not meet pneumonia criteria. 04 among those 39 patients, who had significant colony count of colonizers on BAL culture, were further retrospectively confirmed to actually harbour coloniser, even with BAL procedure as these patients did not respond to antibiotic treatment on follow up. Thus, contingency table was made showing sensitivity of 76.4%, specificity of 89.7%, positive predictive value 90.6% and negative predicative value 74.4% in BAL (Table 2).
Table (2):
Contingency table – BAL Quantitative culture with LRTI {Total microbial isolation= 39(43.3%)}.
| LRTI£ | Total | |||
|---|---|---|---|---|
| Present | Abcent | |||
| BAL | >105 CFU/ml | 39 | 4 | 43 |
| <105CFU/ml | 12 | 35 | 47 | |
| Total | 51 (56.6%) | 39 (43.4%) | 90 | |
£Lower Respiratory Tract Infection
Sensitivity= 39/51=76.4%
Specificity=35/39=89.7%
Positive predictive value=39/43=90.6%
Negative predictive value=35/47=74.4%
On microscopy, Gram stain could accurately identify bacteria in 13 (14.4%) samples; ZN stain for MTB in 08 (8.8%) and KOH mount for fungi in 01 (1.1%) sample. Bacterial growth was seen in 23 (25.5%) samples. CBNAAT was positive in 15 (16.6%) samples and Aspergillus flavus grew in a single sample (1.1%). Bacterial isolation was better in BAL when compared to mycobacteria and fungi as they usually contributed for majority of lower respiratory tract infections (LRTI). Out of 90 samples, 23 (25.5%) samples showed bacterial growth which was in significant colony count (>105CFU/ml) with monomicrobial growth in 19 samples (19 isolates) and polymicrobial growth in 04 samples (08 isolates) resulting in total 27 bacterial isolates. 49 (54.4%) samples showed no growth/ mixed growth/no pathogenic growth/ significant growth (>105CFU/ml) with colonisers, whereas 18 (20%) samples had insignificant colony count. Insignificant growth or coloniser with significant colony count was not a useful indicator in diagnosing LRTI in the present study. Thus, BAL could accurately aid in diagnosing bacterial pneumonia in this study for up to 25.5%, MTB in 16.6% and fungal aetiology in 1.1% with total microbial recovery rate of 39 (43.3%) (Table 3).
Table (3):
Distribution pattern of microorganisms on microscopy and culture from BAL fluid (n=90) Total recovery = 39(43.3%).
Bacteria |
Mycobacteria |
Fungi |
TOTAL |
|
|---|---|---|---|---|
Microscopy |
13 (14.4%) (Gram stain) |
8 (8.8%) (ZN Stain) |
1 (1.1%) (KOH mount) |
22 (24.4%) |
Culture/CBNAAT ϯ |
23 (25.5%) Culture |
15 (16.6%) CBNAAT |
1 (1.1%) Culture |
39 (43.3%) |
ϯ Cartridge based nucleic acid amplification test.
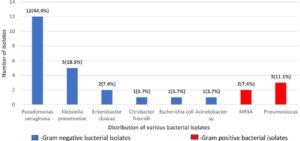
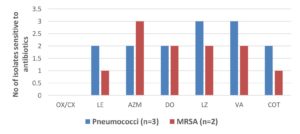
Distribution of Gram positive and Gram-negative isolates is shown in Fig. 1. Of total 27 isolates, 22 (81.5%) were Gram negative isolates and 05 (18.5%) were Gram positive. Amongst Gram negative isolates, Pseudomonas aeruginosa 12 (44.40%) and Klebsiella pneumoniae 05 (18.5%) were isolated predominantly in BAL samples, followed by Enterobacter cloacae complex 02 (7.40%), Citrobacter freundii 01 (3.7%), Escherichia coli 01 (3.7%) and Acinetobacter spp 01 (3.7%). Among Gram positive isolates, Pneumococci 03 (11.10%) and MRSA 02 (7.40%) cases were found. Antibiotic susceptibility pattern of gram-positive isolates is shown in Fig. 2. All 3 isolates of pneumococci were resistant to penicillin which was checked by Oxacillin disc and Penicillin-Minimum inhibitory concentration (MIC) E test. Only 01 (33.3%) among those was MDR. Both MRSA strains isolated were sensitive to vancomycin (by MIC-E test), linezolid, and ciprofloxacin. Thus, a total of 03 (11.1%) Gram positive MDR s were recovered.
Fig. 2. Antibiotic sensitivity pattern of Gram-positive isolates from BAL fluid(n=5).
OX-Oxacillin; CX=Cefoxitin: LE=Levofloxacin: AZM=Azithromycin; DO= Doxycycline; LZ=Linezolid; VA=Vancomycin: COT=Cotrimoxazole
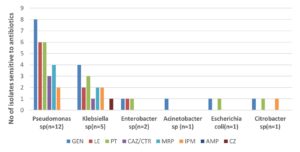
Antibiotic sensitivity pattern in Gram negative isolates varied. 50% (6/12) isolates of Pseudomonas aeruginosa were MDR, whereas 66.7% (8/12) samples showed Carbapenem resistance. 60% (3/5) of Klebsiella pneumoniae were MDR. Single isolate of Acinetobacter spp was MDR with sensitivity to gentamicin and ampicillin sulbactam. Enterobacter cloacae complex (1 isolate) and single isolates of Escherichia coli and Citrobacter freundii were MDRs. Many Gram-negative isolates showed carbapenem and cephalosporin resistance except one Klebsiella pneumoniae which was sensitive to ceftriaxone (Fig. 3). MDR Gram negative bacteria isolated were 13 (48.1%). Thus, a total of 16 (59.2%) MDRs were isolated from 27 bacterial isolates. Rifampicin resistance was detected in 26.6% (4/15) samples by CBNAAT for MTB. Antifungal susceptibility testing for Aspergillus could not be evaluated.
Fig. 3. Antibiotic sensitivity pattern of Gram-negative isolates from BAL fluid(n=22). PT= Piperacillin-Tazobactum: CAZ=Ceftazidime: CTR=Ceftriaxone: MRP=Meropenem: IPM= Imipenem: AMP= Ampicillin: CZ=Cefazolin.
Outpatients without MDR bacteria in BAL were treated as per sensitivity reports. Patients who had MDR bacterial isolation were admitted in view of associated comorbid conditions and presumed morbidity. Renal function tests were done. Two patients with MRSA pneumonia recovered with linezolid therapy for 7-10 days. Single patient with MDR pneumococcal pneumonia recovered with levofloxacin for 7-10 days. Six (6) patients, positive for MDR Gram negative Enterobacterales, were treated with meropenem/ceftazidime-avibactum/colistin/gentamicin, in whom 03 recovered and 03 were lost to follow up. 07 patients with MDR Pseudomonas aeruginosa and MDR Acinetobacter were treated with aminoglycoside/colistin, out of which, 05 cases recovered and 02 were lost to follow up. Patients diagnosed with MTB were treated as per RNTCP guidelines. Single patient who was diagnosed with pulmonary aspergillosis responded well to oral itraconazole therapy.
LRTI affected predominantly middle-aged men in the present study (Mean age-43 yrs) which was comparable to study done in 2020 by Anisha et al16. This male predominance could be due to more outdoor activity, smoking, cultural conditions existing in our country. Comorbidities like diabetes 23 (25.5%), chronic bronchitis 34 (37.7%), COPD 29 (32.2%) and previous history of MTB 11 (12.2%) led to discrepancy in diagnosing LRTI with sputum samples. Hence BAL had to be performed in these patients to aid the diagnosis.
Scientific literature is scarce with regard to the diagnostic utility of BAL in LRTI. Jimenez et al.17 studied patients with suspected LRTI (CAP) without previous antibacterial treatment, BAL has adequate correlation equal to protected brush specimen (PSB) in diagnosing pneumonia. Sensitivity of BAL in the present study was less compared to the study done in 2019 by Travar et al.18 for diagnosing ventilator associated pneumonia, where sensitivity was 91% but it had lesser specificity of 70% compared to the present study, where sensitivity and specificity were 76.4% and 89.7% respectively.
Overall microbial isolation, in the present study was 43.3%, which was found to be similar with other studies like Veleza et al.19 and Kottmann et al.20, where the positive yield was 51.6% and 55.8% respectively. The comparative slight lesser positivity rate in the present study might be for the fact that the study was done in general population, with less sample size and immunocompetent subjects, whereas other studies quoted above were done in immunocompromised patients. 27 (30%) bacterial isolates grew in the present study which is less compared to a recent study done by Adhikari et al21 in 2021, where 95.3% bacterial isolation rate was seen from BAL samples. Utility of BAL in diagnosing mycobacterial infections with Ziehl-Neelson stain was done by Vivek et al.22 in 2016 with positivity rate equal to above study. CBNAAT being extremely sensitive yielded better diagnosis of TB in 15 (16.6%) cases in the present study. Our study had 1.1% fungal growth from BAL. In the study of Vivek et al22, more fungal isolates were found with a predominance of candida albicans (37%) followed by Aspergillus spp (14.8%). The reason for less fungal growth in the present study could be that all the patients were immunocompetent. BAL has shown to be good indicator of fungal infections with comparison equal to that of bronchial biopsy4.
In studies by Kottman et al.20 and Bari et al.23, predominance of aerobic Gram-negative bacteria over Gram positive bacteria was observed. Klebsiella was also the most common pathogen isolated from studies done by Sarmah et al.24, Lin et al.25 and Singh et al.26. Pseudomonas aeruginosa was the commonest isolate in a study done by Magazine et al27 in 2013. The results of present study were comparable to afore mentioned studies demonstrating gram negative organisms emerging predominantly as cause of LRTI from BAL samples. However, Streptococcus pneumoniae was the most common isolate from BAL, found in the study of Bansal et al.28 and Capoor et al.29 which contrasts with the present study.
The present study correlates with the study done by Magazine et al.27 where 87.9% isolates of P. aeruginosa had resistance to cephalosporins and 75% of the isolates of Acinetobacter spp were resistant to meropenem. The findings of the present study were discordant with those of a retrospective study done by Goel N et al.30, which revealed that only 25.6% of the isolates of Acinetobacter species, 22.8% the isolates of P. aeruginosa and 09% of the isolates of Klebsiella species were resistant to meropenem. This difference could be due to the different set up in which study was carried out. In the present study, patients who developed MDR infection of gram negative/gram positive organisms were outpatients, thus needing a more in- depth analysis to address this issue as treatment options for CAP with MDR Enterobacterales, Pseudomonas aeruginosa and Acinetobacter spp are limited due to lack of appropriate antibiotics31,32. Increasing carbapenem resistance in gram negative isolates from BAL samples has been seen in few studies similar to present study33,34.
Current medical literature suggests that Streptococcus pneumoniae is the most common Gram-positive bacterium causing pneumonia, whether community-acquired or hospital acquired. However, as it is evident from the present study, there was predominance of gram-negative organisms and emergence of MDR organisms from those proportion of patients where sputum sample did not yield any conclusive result. This changing trend of gram-negative predominance further needs to be investigated with large multicentre studies. This trend needs to be kept in mind along with patient clinical profile as MDR organisms are emerging as a cause of LRTI due to improper antimicrobial stewardship even in untreated patients.
Limitations of the study
Atypical bacterial pathogens and viruses could not be isolated due to resource constraints and non-affordability of patients in this region. Furthermore, studies need to be conducted to address this issue of emergence of multidrug resistant organisms in community acquired pneumonias.
BAL, as a diagnostic tool can be used accurately to identify varied infections and their causative organisms with sensitivity pattern. BAL is useful not only in bacterial infections but also in fungal infections. A country like India, where tuberculosis is common, BAL may play a vital role in its diagnosis. Clinical profile of the patient needs to be kept in mind while evaluating LRTI. BAL provides that unique information which helps to switch from empirical to precise evidence-based therapy.
ACKNOWLEDGMENTS
We would like to express our heartfelt thanks to Professor Dr. Satyaprasad (Head, Pulmonology department) for his guidance. We would like to thank RNTCP for providing data and ICMR for their support.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
DI contributed to formulating research hypothesis, collection of samples, interpretation and analysis of data along with manuscript writing. AB was involved data analysis, interpretation and contributed novel ideas in presentation of data. PI supervised and reviewed, edited the manuscript, and designed the figures and tables. PP contributed for sample collection, references gathering and patient communication.
FUNDING
This project was awarded with stipend for author PP for initial 1st 2 months only under ICMR STS project Ref no: 2018-00184.No financial assistance from any agencies was taken for further continuation of the study.
ETHICS STATEMENT
This study was approved by the institutional ethical clearance committee. IEC/IRB No-05/2018.
AVAILABILITY OF DATA
All datasets generated or analysed during this study are included in the manuscript.
- Baughman RP. How I Do Bronchoalveolar Lavage. J Bronchol. 2003;10:309-314.
Crossref - Hyaret R. Pulmonary host defenses. Interaction of respiratory antibodies with pseudomonas aeruginosa and alveolar macrophages. J Immunol. 1974;111:369-380.
- Meyer KC, Raghu G, Baughman RP, et al. An official American Thoracic Society clinical practice guideline: The clinical utility of bronchoalveolar lavage cellular analysis in interstitial lung disease. Am J Respir Crit Care Med. 2012;185(9):1004-1014.
Crossref - Radha S, Afroz T, Prasad S, Ravindra N. Diagnostic utility of bronchoalveolar lavage. J Cyto. 2014;31(3):136-138.
Crossref - Mishra SK, Kattel HP, Acharya J, et al. Recent trend of bacterial aetiology of lower respiratory tract infections in a tertiary care center of Nepal. Int J Infect Microbiol. 2012;1(1):3-8.
Crossref - Rano A, Agusti C, Jimenez P, et al. Pulmonary infiltrates in non-HIV immunocompromised patients: a diagnostic approach using non-invasive and bronchoscopic procedures. Thorax. 2001;56(5):379-87.
Crossref - Kim ES, Kim E-C, Lee S-M, et al. Bacterial Yield from Quantitative Cultures of Bronchoalveolar Lavage Fluid in Patients with Pneumonia on Antimicrobial Therapy. Korean J Intern Med. 2012;27(2):156-162.
Crossref - Bartlett JG, Breiman RF, Mandell LA, File TM. Community-acquired pneumonia in adults: guidelines for management. Clin Infect Dis. 1998;26(4):811-838.
Crossref - Musher DM, Thorner AR. Community-acquired pneumonia. N Engl J Med. 2014;371(17):1619-1628.
Crossref - Syrjala H, Broas M, Suramo I, Ojala A, Lahde S. High-Resolution Computed Tomography for the Diagnosis of Community-Acquired Pneumonia. Clin Infect Dis. 1998;27(2):358-363.
Crossref - Thomson RB. Laboratory diagnosis of respiratory infections. Curr Opin Infect Dis. 1999;12(2):115-119.
Crossref - Bhatia GC, Hemvani N, Chitnis DS. Role of bronchoalveolar lavage in pulmonary infections. Lung India. 2006;23(4):147-150.
Crossref - Miller PR, Meredith JW, Chang MC. Optimal threshold for diagnosis of ventilator-associated pneumonia using bronchoalveolar lavage. J Trauma. 2003;55(2):263-268.
Crossref - Bartlett JG, Dowel SF, Mandell LA, File Jr TM, Musher DM, Fine MJ. Practice guidelines for the management of community-acquired pneumonia in adults. Clin Infect Dis. 2000;31:347-382.
Crossref - Pop-Vicas AE, DAgata EM. The rising influx of multi drug resistant gram-negative bacilli into a tertiary care hospital. Clin Infect Dis. 2005;40(12):1792-1798.
Crossref - Anisha CK, Rajkumar HRV, Reddy AR, Ruturaj MK, Manderwad GP. Spectrum of pathogenic bacteria isolated from the bronchoalveolar lavage in a tertiary care centre at Hyderabad. Indian J Microbiol Res. 2020;7(1):48-50.
Crossref - Jiménez P, Saldías F, Meneses M, Silva ME, Wilson MG, Otth L. Diagnostic fiberoptic bronchoscopy in patients with community-acquired pneumonia. Comparison between bronchoalveolar lavage and telescoping plugged catheter cultures. Chest. 1993;103(4):1023-1027.
Crossref - Travar M, Kovacevic P, Golic D. Predictive Values of Quantitative VS Qualitative Culture of Broncholaveolar Lavage in Diagnosis of Ventilator Associated Pneumonia in Patients on Mechanical Ventilation. Int J Inn Res Med. 2019;4(05):304-308.
Crossref - Veleza L, Correa LT, Maya MA, et al. Diagnostic accuracy of bronchoalveolar lavage samples in immunosuppressed patients with suspected pneumonia: Analysis of a protocol. Respir Med. 2007;101(10):2160-2167.
Crossref - Kottmann RM, Kelly J, Lyda E, et al. Bronchoscopy with bronchoalveolar lavage: determinants of yield and impact on management in immunosuppressed patients. Thorax. 2011;66(9):823.
Crossref - Adhikari S, Regmi RS, Pandey S, et al. Bacterial etiology of bronchoalveolar lavage fluid in tertiary care patients and antibiogram of the isolates. J Inst Sci Tech. 2021;26(1):99-106.
Crossref - Vivek KU, Kumar N. Microbiological profile of bronchoalveolar lavage fluid in patients with chronic respiratory diseases: a tertiary care hospital study. Int J Med Res Rev. 2016; 4(3):330-337.
Crossref - Bari SA, Mustafa M. A Study On Bacterial Isolates From Bronchoalveolar Lavage (Bal) Fluid Obtained From Patients With Pulmonary Infections –In Tertiary Care Hospital, Hyderabad. Int J Recent Sci Res. 2018;9(6) 27531-27535.
- Sarmah N, Sarmah A, Das DK. A Study on the Microbiological Profile of Respiratory Tract Infection (RTI) in Patients Attending Gauhati Medical College & Hospital. Ann Int Med Dent Res. 2016;2(5):11-15.
- Lin SH, Kuo PH, Hsueh PR, Yang PC, Kuo SH. Sputum bacteriology in hospitalized patients with acute exacerbation of chronic obstructive pulmonary disease in Taiwan with an emphasis on Klebsiella pneumonia and Pseudomonas aeruginosa. Respirology. 2007;12(1):81-87.
Crossref - Singh AK, Sen MR, Anupurba S, Bhattacharya P. Antibiotic sensitivity pattern of the bacteria isolated from nosocomial infections in ICU. J Commun Dis. 2002;34(4):257-263.
- Magazine R, Chogtu B, Rao S, Chawla K.Bacterial isolates from the bronchoalveolar lavage fluid of patients with pneumonia not responding to initial antimicrobial therapy. Sahel Med J. 2013;16(3):102-106.
Crossref - Bansal S, Kashyap S, Pal LS, Goel A. Clinical and bacteriological profile of community-acquired pneumonia in Shimla, Himachal Pradesh. Indian J Chest Dis Allied Sci. 2004;46:17-22.
- Capoor MR, Nair D, Aggarwal P, Gupta B. Rapid diagnosis of community-acquired pneumonia using the BacT/Alert 3D system. Braz J Infect Dis. 2006;10(5):352-356.
Crossref - Goel N, Chaudhary U, Aggarwal R, Bala K. Antibiotic sensitivity pattern of gram-negative bacilli isolated from the lower respiratory tract of ventilated patients in the Intensive care unit. Indian J Crit Care Med. 2009;13(3):148-151.
Crossref - Savov E, Kovachka K, Trifonova A, Kioseva E, Strateva T. Treatment Options for Infections Caused by Multidrug-Resistant Gram-Negative Bacteria. Acta microbiologica Bulgarica. 2019;35(1):3-8.
- Cilloniz C, Dominedo, C, Torres A. Multidrug-Resistant Gram-Negative Bacteria in Community-Acquired Pneumonia. Crit Care. 2019;23:79.
Crossref - Sisto A, Ancona F, Meledandri M, et al. Carbapenem non-susceptible Klebsiella pneumonia from Micro network hospitals, Italy, 2009 to 2012. Euro Surveill. 2012;17(33):20247.
- Brahmadathan KN, Gladstone P, Rajendran P. Incidence of carbapenem-resistant nonfermenting gram-negative bacilli from patients with respiratory infections in the intensive care units. Indian J Med Microbiol. 2005;23(3):189-191.
Crossref
© The Author(s) 2021. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.