ISSN: 0973-7510
E-ISSN: 2581-690X
Coronavirus disease (COVID-19), which was due to novel coronavirus was detected in December 2019 in Wuhan, China for the first time and spread rapidly became a global pandemic. This study aimed to predict the potential of macroalgae compounds as SARS-CoV-2 antiviral by inhibiting of ACE2 receptor through in silico approach. Twenty-seven macroalgae compounds were obtained from PubChem (NCBI, USA), while target protein ACE2 receptor was collected from Protein Data Bank (PDB). Then the initial screening study drug-likeness conducted by Lipinski rule of five web server and prediction of bioactive probability carried out by PASS (Prediction of activity spectra for biologically active substances) Online web server. After those compounds were approved by Lipinski’s rule of five and PASS online prediction web server, the blind docking simulation was performed using PyRx 0.8 software to show binding energy value. Molecular interaction analysis was done using BIOVIA Discovery Studio 2016 v16.1.0 and PyMOL v2.4.1 software. There are six macroalgae compounds approved by Lipinski’s rule of five and PASS Online Analysis. The result is that macroalgae compound siphonaxanthin among 27 macroalgae compound showed strong binding energy to bind ACE2 receptor with -8.8 kcal/mol. This study also used the SARS-CoV-2 drugs as positive control: remdesivir, molnupiravir, baricitinib, lopinavir, oseltamivir, and favipiravir. The result shows that siphonaxanthin has lowest binding energy than the common SARS-CoV-2 drug. Macroalgae compounds are predicted to have potential as SARS-CoV-2 antiviral. Thus, extension studies need to investigate by in vitro and in vivo analysis for confirmation the siphonaxanthin’s inhibitory activity in combat SARS-CoV-2.
Antiviral, COVID-19, Macroalgae, Medicine, SARS-CoV-2
Coronavirus disease (COVID-19) was detected in late December 2019 in Wuhan City, Hubei Province, China for the first time and spread rapidly became a global pandemic less than six months.1 Currently in 2022, globally the number of new COVID-19 deaths cases were reported over 370 million.2 COVID-19 was due to novel coronavirus, SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) that is categorized pathogenic viral infection and high transmittable.3 SARS-CoV-2 require an ACE2 (angiotensin converting enzyme 2) human receptor for binding. SARS-CoV-2 infect human host cells by an exopeptidase. The virus surface spike protein cleaves by an exopeptidase. Another SARS-CoV-2 structure glycoprotein spike facilitate to bind host cells.4
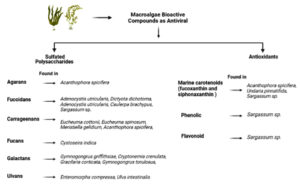
Currently, the therapeutic strategies of antiviral drugs against SARS-CoV-2 were reported, such as baricitinib, lopinavir, remdesivir, and favipiravir.5 Besides that, the application of nanotechnology as SARS-CoV-2 antiviral has emerged promising technologies today.4,6,7 Marine sulphated polysaccharides as one of the most source developed as nanomaterial.8 There are 6 derivative compounds of macroalgae sulfated polysaccharides such as agarans, fucoidans, carrageenans, fucans, galactans, and ulvans
(Fig. 1). Carrageenans one of sulphated polysaccharides derivate which isolated from red macroalgae. Carrageenans have wide spectrum as antiviral, the mechanism by prevent the viral particles when it physical binding happens. Carrageenans were reported combating 12 viruses (SARS-CoV-2, HSV, InfV, hRV, HIV, hCV, hCoV-OC43, HPV, TMV, DENV, JEV, and RVFV).9 Marine algae also produce marine carotenoids such as fucoxanthin and siphonaxanthin compound are rich antioxidants. One of the target compound is needed to combat SARS-CoV-2 is antioxidant.10 It is well known that macroalgae are groups of intertidal organisms that will always be exposed by UV light and extreme conditions. Therefore macroalgae secrete secondary metabolism antioxidants compounds as their self-defense. Sargassum cristaefolium species from brown macroalgae has antioxidant activity, that was total phenolic content 44.95±2.62 mg gallic acid/g extract and 70.27±3.59 g quercetin/g extract was total flavonoid content.11 Siphonaxanthin compounds from Codium fragiles have potential as antiviral with an activity IC50 = 87.4 μM.12
The recent in silico study related have screened 12 algae compounds by inhibit ACE2 receptor, the result showing that 4 compounds have good score.13 This study is important for more exploring and focusing on macroalgae derivative compounds such as carotenoids and sulfated polysaccharides as SARS-CoV-2 antiviral by molecular mechanism reasoning. This study aimed to predict the potential of macroalgae compounds as SARS-CoV-2 antiviral by inhibit ACE2 receptor with in silico study approach, that are capable to predict the cellular pathways, molecular interactions type, and binding energy of candidate compound.14 Besides that, the important of an in silico approach as primarily prediction before doing in vitro or in vivo study. In silico research can assist identify promising compounds for medication creation and widespread exploitation of secondary metabolites from natural resources.
Materials
The materials in this study were done by hp computer hardware, the specifications are processor Intel® CORETM i5-DDR4-3200 MHz RAM. While, the software be used to molecular carried out by PyRx v.0.8 (Scripps Research, USA),15 PyMOL v.2.4.16 (Schrodinger Inc, USA) for protein sterilization and visualization of the interaction of ligand-target protein, and DS BIOVIA Discovery Studio 2016 v16.1.0 x64 (Dassault Systèmes, France) for analysis of the docking simulation.17 This study also used the Lipinski rule of five (http://www.scfbio-iitd.res.in/software/drugdesign/lipinski.jsp) and PASS Online web server (http://way2drug.com/passonline/) for primary study.
Ligand and Target Protein Preparation
The carotenoids derivate of macroalgae and sulfated polysaccharides was collected from PubChem (https://pubchem.ncbi.nlm.nih.gov/), 27 ligand compounds were collected (Table 1). Protein Data Bank (https://www.rcsb.org/) as source for ACE2 target protein collected, then prepared by PyMOL v.2.4.1 (Schrödinger Inc, USA).
Initial Screening Study
Lipinski’s rule of five and PASS online prediction web server conducted in this step. There are five rules of Lipinski’s rules as like nether than 500 dalton of molecular weight, nether than 5 the number of donor hydrogen bonds, nether than 10 the number of acceptor hydrogen bonds, and nether than 5 of high lipophilicity, two rules from five rules are littlest requirements.18 This step aims for analyzing the drug-likeness from macroalgae compounds. While for prediction the candidate compound potential of activation (Pa) and inhibition (Pi) was done by PASS online prediction. Ideally, Pi value must be lower than Pa value.
Molecular Docking Study
After 27 ligand compounds were done by initial screening study, then molecular docking study by PyRx 0.8 version15 for estimating binding energy value of ligand and ACE2 receptor interaction. Blind docking was used to predict ligand bind the target protein ACE2 receptor.
Visualization of Protein-Ligand Interaction
To analysis the protein and ligand interaction was conducted by DS BIOVIA Discovery Studio 2016 v16.1.0 x64 (Dassault Systèmes, France) and PyMOL v.2.4.1 (Schrödinger Inc, USA) software. This analysis for prediction the type of chemical bond and interaction position when macroalgae compounds bind ACE2 receptor.
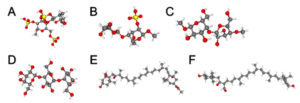
In this in silico study aimed for screening macroalgae compounds potential as ACE2 inhibitor. The Macroalgae derivative compounds such as carotenoids and sulphated polysaccharides. Macroalgae compounds were collected from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/). There were 27 compounds collected (Table 1), then an initial screening study was conducted by Lipinski rule of five web server (http://www.scfbio-iitd.res.in/software/drugdesign/lipinski.jsp), the result is 25 compounds was approved by Lipinski rule of five and then 25 compounds carried out by PASS online prediction (http://www.way2drug.com/passonline/) web server. The result showed that 6 compounds were approved that have the potential as antiviral. The 3D structure of 6 compounds was approved shown in Fig. 1.
Table (1):
The ligand compounds were collected.
No. |
Compound |
PubChem CID |
|---|---|---|
1. |
An Agaran |
405237310 |
2. |
Carrageenan |
71597331 |
3. |
kappa-Carrageenan |
11966249 |
4. |
lambda-Carrageenan |
101231953 |
5. |
lambda-Carrageenan (High-viscosity) |
146680192 |
6. |
kappa-Carrageenan; 11114-20-8 |
73155740 |
7. |
Carrageenan; 9000-07-1 |
78126884 |
8. |
lambda-Carrageenan |
91972149 |
9. |
beta-Carrageenan |
102199626 |
10. |
alpha-Carrageenan |
102199625 |
11. |
ZINC acetate |
11192 |
12. |
Fucoidan; 9072-19-9 |
402346915 |
13. |
Fucoidan; C08253; 9072-19-9 |
10452 |
14. |
Fucoidan; W-204037 |
255374929 |
15. |
A fucan with alpha-(1->3) bonds |
405236660 |
16. |
Fucoidan; sulfated Fucose; Sulfated Alga Polysaccharide; NSC631568; |
596143 |
17. |
Fucoidan; SC-16162; 9072-19-9 |
335958879 |
18. |
A fucan with alpha-(1->4) bonds |
405234669 |
19. |
Fucoidan; 9072-19-9 |
406854333 |
20. |
Galactan |
53477780 |
21. |
Quinidine Arabino Galactan Sulfate |
76519688 |
22. |
(1->4)-beta-galactan |
53356679 |
23. |
ulvan |
405234592 |
24. |
a fucan with alternating alpha-(1->3) and alpha-(1->4) bonds |
405234857 |
25. |
a fucan with alternating alpha-(1->4) bonds |
405234669 |
26. |
fucoxanthin |
5281239 |
27. |
siphonaxanthin |
11204185 |
Table (2):
The result of Lipinski rule of five.
| Compound | Lipinski Rule of Five | ||||
|---|---|---|---|---|---|
| MW (Dalton) | HBD | HBA | LOGP | MR (g/mol) | |
| Lambda-Carrageenan | 594.000 | 3 | 19 | 1.297 | 109.809 |
| Alpha-Carrageenan | 416.000 | 3 | 12 | 1.719 | 85.746 |
| Galactan | 370.000 | 6 | 11 | 1.754 | 82.579 |
| (1->4)-beta-galactan | 504.000 | 10 | 16 | 1.457 | 106.314 |
| Fucoxanthin | 312.000 | 5 | 6 | -0.053 | 77.145 |
| Siphonaxanthin | 312.000 | 5 | 6 | -0.053 | 77.145 |
Note:MW: Molecular Weight, HBD; Hydrogen Bond Donors, HBA; Hydrogen Bond Acceptors, LOGP; High Lipophilicity, MR; Molar Refractivity.
Table 2 present the results of Lipinski’s five rule which generally predicts a compound as a drug like molecule. Druglikeness explained as the complex balance of diverse molecular properties and structural features that determine whether a particular molecule categorized as a drug or not. That properties include molecular weight, characteristics of hydrogen bonding lipophilicity, and existence of various pharmacophoric features in that compounds.19 The result is molecular weight value of alpha-carrageenan, galactan, fucoxanthin, and siphonaxanthin under 500.000 dalton, it is predicted that alpha-carrageenan, galactan, fucoxanthin, and siphonaxanthin can enter the cell membrane. While the molecular weight of lambda-carrageenan and (1->4)-beta-galactan have a molecular weight more than 500.000 dalton, so that both of the compound estimated unable to pass membrane cells. Advanced study be required for making molecular weight smaller under 500.000 dalton. Log P value indicates the coefficient of solvability in water or fat, the coefficient range is -0.4 to 5. The larger log P number represents that the more hydrophobic molecules characteristics would be. The over hydrophobic molecules cause the increased level of toxicity effect because of molecules longer disconnected in the lipid bilayer membrane will be spread broadly in the body. Thus, the selective effect of the compound inhibit target protein is reduced. Meanwhile, the more negative the log P number, the more non-permeable the molecule will be.20,21
Table (3):
The result of PASS online prediction.
Compound |
Activity |
Pa |
Pi |
|---|---|---|---|
Lambda-Carrageenan |
Antiviral |
0.654 |
0.004 |
Alpha-Carrageenan |
Antiviral |
0.656 |
0.004 |
Galactan |
Antiviral |
0.735 |
0.004 |
(1->4)-beta-galactan |
Antiviral |
0.684 |
0.007 |
Fucoxanthin |
Antiviral |
0.244 |
0.135 |
Siphonaxanthin |
Antiviral |
0.393 |
0.099 |
Table 3 shows the result of PASS online prediction webserver for prediction the ability of the bioactive compound from macroalgae as antiviral for activation or inhibition by human cells. The principle is that the Pa (potential activation) value must be higher than Pi (potential inhibition) value. In this in silico study Pa>0.3. The results showed macroalgae Pa value larger than its Pi value. It is indicates when a macroalgae compound penetrate human body, it have potential to inhibit ACE2 protein.
Table (4):
The result of molecular docking simulation.
Compound |
PubChem CID |
Target Protein |
Binding Energy (kcal/mol) |
|---|---|---|---|
Siphonaxanthin |
11204185 |
ACE2 |
-8.8 |
Fucoxanthin |
5281239 |
ACE2 |
-8.7 |
(1->4)-beta-galactan |
53356679 |
ACE2 |
-8.3 |
Alpha-Carrageenan |
102199625 |
ACE2 |
-7.5 |
Lambda-Carrageenan |
101231953 |
ACE2 |
-7.4 |
Galactan |
53477780 |
ACE2 |
-7.0 |
Table 4 shows the result of macroalgae compound molecular docking simulation. Molecular docking simulation was done by PyRx 0.8 version and the followed grid docking result are center X: 166.4430 Y: 223.6339 Z: 301.4075 and dimensions (Å) X: 67.1115 Y: 74.6594 Z: 67.3432. Due to the target protein functional domain is unknown, so in this study aimed blind docking method.22
Table (5):
The result of molecular docking simulation from drugs as control.
Drug as Control |
PubChem CID |
Target Protein |
Binding Energy (kcal/mol) |
|---|---|---|---|
Remdesivir |
121304016 |
ACE2 |
-7.7 |
Molnupiravir |
145996610 |
ACE2 |
-7.5 |
Baricitinib |
44205240 |
ACE2 |
-7.1 |
Lopinavir |
92727 |
ACE2 |
-6.8 |
Oseltamivir |
65028 |
ACE2 |
-6.2 |
Favipiravir (Avigan) |
492405 |
ACE2 |
-5.7 |
Based on the result of the binding energy value from the macroalgae compound selected, siphonaxanthin has the lowest binding energy to bind ACE2 receptor with binding energy value -8.8 kcal/mol. While, from sulfated polysaccharides derivate compound (1->4)-beta-galactan has the lowest binding energy with -8.3 kcal/mol. Ligands with the lowest binding energy are predicted to have a target protein’s biological activity. It refers to the aim of this study the compound is predicted to have the ability to inhibit ACE2 receptor. The lowest binding energy interaction ligand-target protein allows molecular complex formation constant temperature and pressure.23 This study also using common antiviral drug such as remdesivir, molnupiravir, baricitinib, lopinavir, oseltamivir, and favipiravir (avigan).5,24-26 These antiviral common drugs used as positive control for comparing the binding energy value to target protein ACE2 receptor. Based on the binding energy result, siphonaxanthin, fucoxanthin, (1->4)-beta-galactan, and (1->4)-beta-galactan has the lowest binding energy than antiviral common drug.
Fig. 2. 3D structure of macroalgae chemical compound from the PubChem database. Lambda carrageenan (CID: 101231953) (A), alpha carrageenan (CID: 102199625) (B), galactan (CID: 53477780) (C), (1->4)-beta-galactan (CID: 53356679) (D), fucoxanthin (CID: 5281239) (E), and siphonaxanthin (CID: 11204185) (F).
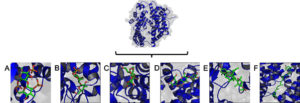
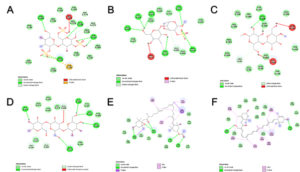
The docking simulation is finished, then molecular complex visualization was conducted by PyMOL v.2.4.1 showed in Fig. 2. Macroalgae compounds can bind to the ACE2 receptor. Fig. 3 present the complex ligands-protein visualization 2D diagram was carried out by DS BIOVIA Discovery Studio 2016 v16.1.0 x64. There are various bond types of macroalgae compounds with ACE2 receptor. Macroalgae carotenoids such as fucoxanthin and siphonaxanthin showed that Van der Waals, Alkyl, and Pi-Alkyl formed the dominant type bond (Fig. 4 and Table 6). While in Macroalgae sulfated polysaccharides, the dominant type bond formed is Van der Waals and Conventional Hydrogen Bond. The higher hydrogen bonds total between protein-ligand complex, the more stable the interaction of protein-ligand.21
Table (6):
The summary of amino acid residues from macroalgae.
Compound |
Amino Acid Residues |
Bond Type |
|---|---|---|
Lambda-Carrageenan |
SER A:511, GLU A:398, THR A:347, HIS A:345, GLU A:402, PRO A:346, HIS A:378, TYR A:381, ASP A:382, TRP A:349, PHE A:40,ARG A:514, TYR A:515, ALA A:348, ASP 350, GLU A:375, HIS 374,HIS A:401,TYR A: 510 |
Van der Waals, Conventional Hydrogen Bond, Carbon Hydrogen Bond, Unfavorable Donor-Donor, Pi-Sulfur |
Alpha- Carrageenan |
THR A:347, GLU A:406, THR A:445, HIS A:374, PRO A:346, HIS A:345,TYR A:515, ARG A:518, THR A:371, GLU A:375, HIS A:378, ALA 348,GLU A:402,ARG A:514, TYR A:515 |
Van der Waals, Conventional Hydrogen Bond, Carbon Hydrogen Bond, Unfavorable Donor-Donor, Pi-Alkyl |
Galactan |
GLU A:398, TYR A:207, TRP A:566, ALA A:396, VAL A:209, PRO A:565, GLU A:564, ALA A:99, GLN A:98, GLY A:205, TYR A:196, TYR A:202, ASP A:206, GLU A:208,LEU A:95,LYS A:562, ASN A:394 |
Van der Waals, ConventionalHydrogen Bond, Carbon Hydrogen Bond, Unfavorable Donor-Donor |
(1->4)-beta-galactan |
TYR A:515, TRP A:349, TYR A:385, TYR A:381, HIS A:378, THR A:347, PRO A:346, GLU A:375, HIS A:374, ARG A:514, ASP A:350, HIS A:401, ASP A:382,HIS A:345, GLU A:402, ALA 348,ALA A:348 |
Van der Waals, Conventional Hydrogen Bond, Carbon Hydrogen Bond, Unfavorable Donor-Donor |
Fucoxanthin |
THR A:347, GLU A:375, PRO A:346, ALA A:348, HIS A:401, GLU A:402, ASN A:508, THR A:125, TYR A:127, GLU A:145, HIS A:378,ARG A:514, SER A:124, SER 128,PHE A:504, HIS A:378, PHE A:504, HIS A:345, TYR A:510 |
Van der Waals, Conventional Hydrogen Bond, Pi-Sigma, Alkyl, Pi-Alkyl |
Siphonaxanthin |
ARG A:518, TYR A:515, HIS A:345, ARG A:514, ASP A:509, LYS A:187, MET A:190, GLU A:398, SER A:511, THR: A:445, ASP A:367, LEU A:370, GLU A:406, THR A:371, GLU A:402, TYR A:202,PRO A:346, HIS A:374,HIS A:378, HIS A:505, PHE A:504, TYR A;510, TRP A:203 |
Van der Waals, Conventional Hydrogen Bond, Alkyl, Pi-Alkyl |
Fig. 3. Molecular visualization of macroalgae bind to ACE2 protein receptor. The ACE2 protein receptor displayed on transparant surface and blue cartoon structure.Lambda carrageenan (CID: 101231953) (A), alpha carrageenan (CID: 102199625) (B), galactan (CID: 53477780) (C), (1->4)-beta-galactan (D) (CID: 53356679), fucoxanthin (CID: 5281239) (E), and siphonaxanthin (CID: 11204185) (F).
Fig. 4. Chemical interaction between macroalgae compound with target protein ACE2 receptor. Lambda carrageenan (CID: 101231953) (A), alpha carrageenan (CID: 102199625) (B), galactan (CID: 53477780) (C), (1->4)-beta-galactan (D) (CID: 53356679), fucoxanthin (CID: 5281239) (E), and siphonaxanthin (CID: 11204185) (F).
There are various types of amino acid residues that form an interaction between ligand and target protein ACE2 receptor, shown in Table 5. Binding to these amino acid residues could potentially contribute to structural alterations of the ACE2 receptor, resulting in functional changes. Therefore, futher research related to in vitro and in vivo study will be interest to know the mechanism of macroalgae compounds as ACE2 receptor and against SARS-CoV-2.
Nowadays, in silico studies is important for drug development against SARS-CoV-2. Recent studies promising macroalgae compounds as SARS-CoV-2 drug candidates such as sulfated polysaccharides can inhibit SARS-CoV-2 by binding RBD spike protein. There are 17 seaweed sulphated polysaccharides that were screened as SARS-CoV-2 antiviral. The docking results lowest binding energy is -8.2 kcal/mol from xylan sulphate with positive control using heparin tetrasaccharide N-sulfated and heparin.27 In comparison, the other in silico study used 9 investigated compounds marine sulphated polysaccharides promising antiviral agents and using heparin as positive control.28 In both studies used heparin as the positive control. Meanwhile, other studies recommend remdesivir, molnupiravir, baricitinib, lopinavir, oseltamivir, and favipiravir as drugs against SARS-CoV-2. Hence, it is needed for extensive research to develop SARS-CoV-2 antiviral drugs using another positive drug control. Both of those studies do not explain the drug-likeness analysis Lipinski rule of five and predict the bioactive compounds as antiviral using PASS online prediction webserver. Hence, this research is important to conduct with initial screening studies that were done by Lipinski rule of five and PASS online prediction webserver to screen sulfated polysaccharides as antiviral drugs. Other than that, macroalgae is rich with antioxidant compounds that have the potential as SARS-CoV-2 antiviral. Therefore, this study also conducted molecular docking to screen macroalgae antioxidant compounds as SARS-CoV-2 antiviral.
The potential of macroalgae as SARS-CoV-2 antiviral is predicted by the inhibition of ACE2 receptor. By the Lipinski rules of five and PASS online prediction result there are 6 macroalgae compounds were approved from 27 macroalgae collected from PubChem database. Molecular docking results show compound that has lowest binding energy is siphonaxanthin. Siphonaxanthin has the lowest binding energy than the SARS-CoV-2 common drug. Hence, in vitro and in vivo study related to confirm the inhibitory activity of siphonaxanthin against SARS-CoV-2.
ACKNOWLEDGMENTS
The authors would like to thank Generasi Biologi Indonesia Foundation, Indonesia and the Bioscience and Biotechnology Research Centre, Mataram University, Indonesia.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
HP, VDK, ANMA, MHW, MEU drafted the manuscript, compiled information from the literature, and designed the Figures and tables. MTS, OG, SC, NB, ESP, THS, RZ supervised the study. VDK, ANMA, THS, RZ supervised and reviewed the manuscript. All authors read and approved the final manuscript for publication.
FUNDING
None.
ETHICS STATEMENT
Not applicable.
AVAILABILITY OF DATA
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
- Machhi J, Herskovitz J, Senan AM, et al. The natural history, pathobiology, and clinical manifestations of SARS-CoV-2 infections. J Neuroimmune Pharmacol. 2020;15(3): 359-386.
Crossref - Dong E, Du H, Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis. 2020;20(5):533-534.
Crossref - Liu YC, Kuo RL, Shih SR. COVID-19: The first documented coronavirus pandemic in history. Biomed J. 2020;43(4):328-333.
Crossref - Sarangi MK, Padhi S, Dheeman S, et al. Diagnosis, prevention, and treatment of coronavirus disease: A review. Expert Rev Anti Infect Ther. 2022;20(2):243-266.
Crossref - Frediansyah A, Tiwari R, Sharun K, Dhama K, Hapan H. Antivirals for COVID-19: A critical review. Clin Epidemiol Glob Gealth. 2021;9:90-98.
Crossref - Chakravarty M, Vora A. Nanotechnology-based antiviral therapeutics. Drug Deliver Trans Res. 2021;11(3):748-787.
Crossref - Weiss C, Carriere M, Fusco L, et al. Toward Nanotechnology-Enabled Approaches against the COVID-19 Pandemic. Acs Nano. 2020;14(6):6383-6406.
Crossref - Manivasagan P, Oh J. Marine polaccharide-based nanomaterials as a novel source of nanobiotechnological applications. Int J Biol Macromol. 2016;82:315-327.
Crossref - Salih AEM, Thissera B, Yaseen M, et al. Marine sulfated polysaccharides as promising antiviral agents: A comprehensive report and modeling study focusing on SARS CoV-2. Mar Drugs. 2021;19(8):406.
Crossref - Choudhary J, Dheeman S, Sharma V, et al. Insights of severe acute respiratory syndrome coronavirus (SARS-CoV-2) pandemic: A current review. Biol Proced Online. 2021;23(1):5.
Crossref - Prasedya ES, Frediansyah A, Martyasari NWR, et al. Effect of particle size on phytochemical composition and antioxidant properties of Sargassum cristaefolium ethanol extract. Sci Rep. 2021;11(1):17876.
Crossref - Yim SK, Kim I, Warren B, Kim J, Jung K, Ku B. Antiviral activity of two marine carotenoids against SARS-CoV-2 virus entry in silico and in vitro. Int J Cell Sci Mol Biol. 2021;22(12):6481.
Crossref - Rauf A, Rashid U, Khalil AA, et al. Docking-based virtual screening and identification of potential COVID-19 main protease inhibitors from brown algae. S Afr J Bot. 2021;143:428-434.
Crossref - Kharisma VD, Widyananda MH, Ansori ANM, Nege AS, Naw SW, Nugraha AP. Tea catechin as antiviral agent via apoptosis agonist and triple inhibitor mechanism against HIV-1 infection: A bioinformatics approach. J Pharm Pharmacogn Res. 2021;9(4):435-445.
- Kharisma VD, Agatha A, Ansori ANM, et al. Herbal combination from Moringa oleifera Lam. and Curcuma longa L. as SARS-CoV-2 antiviral via dual inhibitor pathway: A viroinformatics approach. J Pharm Pharmacogn Res. 2021;10(1):138-146.
- Kharisma VD, Ansori ANM, Nugraha AP. Computational study of ginger (Zingiber officinale) as E6 inhibitor in human papillomavirus type 16 (HPV-16) infection. Biochem Cell Arch. 2020;20(Suppl 1):3155-3159.
Crossref - Yadav S, Pandey SK, Singh VK, Goel Y, Kumar A, Singh SM. Molecular docking studies of 3-bromopyruvate and its derivatives to metabolic regulatory enzymes: Implication in designing of novel anticancer therapeutic strategies. PLoS One. 2017;12(5):e0176403.
Crossref - Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001;40(1-3):3-26.
Crossref - Ansori ANM, Kharisma VD, Parikesit AA, et al. Bioactive compounds from mangosteen (Garcinia mangostana L.) as an antiviral agent via dual inhibitor mechanism against SARS-CoV- 2: An in silico approach. Phcog J. 2022;14(1):85-90.
Crossref - Shi H, Cui Y, Qin Y. Discovery and characterization of a novel tryptophan hydroxylase 1 inhibitor as a prodrug. Chem Biol Drug Des. 2018;91(1):202-212.
Crossref - de Freitas RF, Schapira M. A systematic analysis of atomic protein-ligandinteractions in the PDB. Med Chem Comm. 2017;8(10):1970-1981.
Crossref - Hassan NM, Alhossary AA, Mu Y, Kwoh CK. Protein-ligand blind docking using QuickVina-W with inter-process spatio-temporal integration. Sci Rep. 2017;7(1):1-13.
Crossref - Ramirez D, Caballero J. Is it reliable to use common molecular docking methods for comparing the binding affinities of enantiomer pairs for their protein target? Int J Mol Sci. 2016;17(4):525.
Crossref - Frediansyah A, Nainu F, Dhama K, Mudatsir M, Harapan H. Remdesivir and its antiviral activity against COVID-19: A systematic review. Clin Epidemiol Glob Gealth. 2021;9:123-127.
Crossref - Cao B, Wang Y, Wen D, et al. A trial of lopinavir-ritonavir in adults hospitalized with severe Covid-19. N Engl J Med. 2020;7:1787-1799.
Crossref - Shim SJ, Chan M, Owens L, Jaffe A, Prentice B, Homaira, N. Rate of use and effectiveness of oseltamivir in the treatment of influenza illness in high-risk populations: A systematic review and meta-analysis. Health Sci Rep. 2021;4(1):e241.
Crossref - Douma M, Boualy B, Manaut N, et al. Sulphated polysaccharides from seaweeds as potential entry inhibitors and vaccine adjuvants against SARS-CoV-2 RBD spike protein: A computational approach. J Taibah Univ Sci. 2021;15(1):649-655.
Crossref - Hans N, Malik A, Naik S. Antiviral activity of sulfated polysaccharides from marine algae and its application in combating COVID-19: Mini review. Bioresour Technol Rep. 2021;13:100623.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.