ISSN: 0973-7510
E-ISSN: 2581-690X
Effluents containing dyes from different industrial sectors pose a serious threat to the environment. Different physicochemical strategies are being carried out in industry to reduce the toxicity of dye-containing waste so that dye-mixed wastewater can be further utilized in agriculture or irrigation purposes in water-scarce areas. But those techniques are economically not feasible. There is an alternative mechanism present in biological systems that are biocatalysts which is eco-friendly, low cost, and sustainable. Lignin peroxidase, Laccase, Manganese peroxidase are oxidoreductase classes of enzymes with the ligninolytic ability and are potential biocatalysts for the degradation of environmental toxicants like dyes. Besides ligninolytic enzymes, cellulase, pectinase are also powerful candidates for dye decolourization. Most interestingly these biocatalysts are found in a variety of microbial monoculture as well as in mixed microbial consortia. The consortia are able to reduce the organic load of dye-containing industrial effluent at a higher rate rather than the monoculture. This article critically reviews the efficacy of lignocellulolytic enzymes in dye decolourization by both monoculture and consortia approaches. In addition, this review discusses the genetically and metabolically engineered microbial systems that contribute to dye decolourization as well as put forward some future approaches for the enhancement of dye removal efficacy.
Dye Decolourization, Lignocellulolytic Enzymes, Microbial Consortia, Genetic Engineering
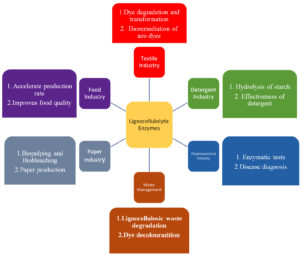
Globally, large-scale synthetic polymers are manufactured from fossil resources. However, our fossil resources are depleting, and their usage has fatal environmental consequences, necessitating a continuous search for alternate and viable sources that can replace fossil fuels while still delivering useful end goods.1 Lignocellulosic biomass is an alternative and is derived from agricultural crops and forest leftovers, solid municipal wastes, as well as paper-mill sludge, bioenergy crops, animal manures, and forest products. Dyes are coloured chemicals that are commonly used in the printing, textile, rubber, plastics, cosmetics, and leather sectors. In our daily lives, natural and synthetic dyes play an important role. Natural dyes like haematein and hematoxylin are derived from logwood that can be toxic if inhaled, swallowed, or absorbed via the skin. When breathed, bloodroot, the source of another natural dye, can cause irritation and inflammation.2 Heavy metals with colours and cancer-causing synthetic dyes have a high impact and are unacceptably harmful. Synthetic dyes can cause an allergic reaction, respiratory trouble, and skin sensitization in industry workers.3 In humans, synthetic cationic dyes can cause hypertension, shock, vomiting, cyanosis, jaundice, quadriplegia, Heinz body development, and tissue necrosis. Therefore, decolourization of the dye is important for lowering its organic load. There are several dye decolourization processes. Among them,the lignocellulolytic enzyme is one of the best sustainable routes. These enzymes are obtained from microorganisms distributed in both prokaryotic and eukaryotic domains including bacteria, fungi, and actinomycetes. Biological pretreatment has various advantages, including minimal energy consumption, the absence of hazardous chemicals, and reduced pollution. Lignocellulolytic enzymes can be characterized as a large group of extracellular proteins, which include hydrolytic activity such as laccase, lignin peroxidase, hemicellulases, cellulases, pectinase, amylase, chitinases, proteases, esterases, mannanases, which are capable of digesting rigid lignocellulose in plant biomass. Lignin and polysaccharides such as cellulose, hemicellulose, pectin, ash, minerals, and salts make up lignocellulosic biomass. Lignin is an aromatic polymer, unlike cellulose and hemicellulose, which are both carbohydrates. Lignocellulose is a valuable source of renewable carbon that has been largely underutilized. Pretreatment of recalcitrant lignocellulosic biomass for biofuel generation, use in the paper, textile, and food industries, wastewater treatment, bioremediation, organic synthesis, and the cosmetic and pharmaceutical sectors are all examples of lignin-degrading enzyme applications. The peroxidases like lignin peroxidase (LiP), manganese peroxidase (MnP), versatile peroxidase (VP), and dye-decolourizing peroxidase (DyP) are all known for the breakdown of lignin.4 DyP enzymes are known originally to oxidize anthraquinone dyes with strong redox potential. DyP has a vast substrate affinity that can act at lower pH levels. Laccases are the second most common type of lignin-degrading enzyme. Lignin-degrading enzymes are already being used in industries including paper and textiles, as well as for wastewater treatment and herbicide breakdown.5 In the delignification and bio-bleaching of wood pulp, LiP, MnP, VP, and laccase work to replace chlorine-based delignification. They can also be used to decolourize dye wastewater from the textile industry, as well as decolourize effluent and treat effluent in distilleries and waste treatment facilities. To this end, the aim of this review is to take a look at the role of different types of lignocellulolytic microbial systems used in dye decolourization along with the critical analysis of their efficiencies (Figure 1) with multiple future prospective mechanisms that can become a perfect workhorse for future research aspirants.
Figure 1. Multiple functions of lignocellulolytic enzymes
Here, the use and application of lignocellulolytic enzymes in different industries have been illustrated.
Critical Analysis Of Dye Decolourizing Lignocellulolytic Enzyme Producing Microbial Systems & Its Significance
Here we are going to review some lignocellulolytic enzymes that have dye decolourizing properties.
Laccase
Laccase (1,4-benzenediol) oxidizes several aromatic substances leading to the simultaneous reduction of molecular oxygen to water.6 Laccase can also be classified in respect of three different kinds of copper prosthetic groups. Though phenolic and its relevant compounds act as the main substrate of this enzyme, it is possible to extend the range of substrate by adding specific substances known as mediators. A mediator is a substance that causes the extension of the substrate range of any enzyme.7 Laccases have been reported to be produced by several gram-positive and gram-negative bacteria, fungi, and actinomycetes. Both Bacterial and fungal laccase require the mediator system. There are several mediators like ABTS(2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid)), HOBt(Hydroxybenzotriazole), and syringaldehyde which lead to the enhancement of redox potential of prokaryotic laccases.8 Nowadays, laccase has been the center of attraction in research fields because of its wide range of applications such as textile dyes bleaching, detoxification of effluent and pollutants, bioremediation of phenolic compounds.9 Dye effluents cause harmful impacts on the soil and groundwater levels. Laccase can play an important role as a biological tool for several applications such as decolourization of dye effluents of industrial textiles.7 At first, the prokaryotic laccase was discovered in the soil bacterium named Azospirillum lipoferum. The melanogenic bacteria Marinomonas mediterranea also has the potential to produce heterologously expressed laccase. CotA gene product, one of the best laccases, is part of the spore coat of Bacillus subtilis.10 Low molecular weight extracellular fungal laccases are produced from Volvariella volvacea11 and Marasmius quercophilus. Extracellular laccases are also found in the bacteria B. safensis, B. lichiniformis,12 B. atrophaeus, B. pumilis,13 B. subtilis, and B. tequileasis. The molecular weight of the laccases from these bacteria ranges from 58kDa to 63kDa. The laccases extracted from fungi exhibit activity in acidic pH. Laccase produced by B. halodurans is considered as alkaline bacterial laccase because it is able to show activity in pH optima 7.5-8.0 in presence of substrate i.e., syringaldazine. Some of the bacterial laccases have the potential to demonstrate activity and stability in a wider pH range i.e., 6.0- 9.0 approximately. In comparison to fungal laccase, bacterial laccases can be applied to numerous industrial fields like bio-bleaching and processing of dyestuffs because of their activity over a wider range of pH. Marine Alteromonas sp. possesses the ability to produce laccase. Synthetic dyes that are released from textile industries pose a huge environmental threat to the human race and the environment due to their carcinogenic properties. Microbial decolourization methods render a less costly and eco-friendly alternative to the current physicochemical process such as adsorption, chemical transformation. Decolourization with the help of laccase is performed on numerous types of dyes that include indigoid, azo, triarylmethane, and anthraquinone dyes. As the current methods are unable to degrade dye mixture while treating wastewater mixed with azo dye, the utilization of a laccase mediated system represents a feasible solution to this alarming issue. The production of laccase can be influenced by the presence of several dye-containing media.7 The first case of degrading dyes has been reported by the laccase from Gamma-proteobacterium JB. The laccase extracted from this microorganism is alkali tolerant and possesses the capability to cause degradation of the dye named indigo carmine at 55°C in pH 9.0. The increase in the rate of breakdown has been found in the presence of syringaldehyde, vanillin, and p-hydroxybenzoic acid. The fungal laccase purified from Trametes hirsuta (pH 0.5) and Sclerotium rolfsii (pH 5.5) had shown results on this dye. Ganoderma lucidum has the potential to cause the production of laccase that contributes to the decolourization of acid orange dye. Streptomyces ipomoeae causes the production of laccase SilA that remains active in neutral to alkaline pH in textile wastewaters.14 It has been checked that the azo dyes are generally degraded by Streptomyces species. But S. psammoticus demonstrates very limited activity towards decolourization. Double mutant K316N/D500G of the Bacillus licheniformis CotA reported in the research by Koschorreck et al. in 2009 exhibits a significant role in the decolourization of dyes such as Remazol Brilliant Blue R (RBBR), Alizarin red S, and indigo carmine (indigo dye).7 Extracted laccase from Streptomyces coelicolour also contains the ability to decolourize the indigo dye in presence of syringaldehyde that acts as a redox mediator.15
Lignin Peroxidase
Lignin is basically a complex aromatic polymer and the most important renewable carbon source on Earth just after cellulose, which consists of about 30% non-fossil organic carbon. Around 60% of lignin is wasted by combustion because of the lack of methods to convert it into valuable end products. For the competitiveness of biofuel vs fossil gasoline, lignin degradation represents the major hindrance. Enzymes like lignin peroxidase (LiP) play an important role in dye – decolourization. Plant cell walls have lignocellulosic biomass as a structural component that mainly comprises cellulose, hemicelluloses, and lignin. In the case of plant lignin-carbohydrate complexes formed with cellulose and hemicelluloses. Wood–decaying fungi, typically classified as white rot-fungi have their extracellular ligninolytic enzymes, basically secreted from fungal class II peroxidase, such as lignin peroxidase (LiP). Lignin has been marked as a complex aromatic structure that is harder to attack and has been considered by most biorefineries to be a low-cost product. Lignin cross-linking is obtained through the oxidative coupling of the various small units such as sinapyl, Coniferyl, and p-coumaryl alcohol. Given the composition of these compounds, lignin is an excellent source of potentially fine chemicals and their evolution through a manufacturing process. Currently, ligninolytic enzymes have many applications like the removal of dye from industrial and bioleaching effluents and the treatment of wastewater. Both phenolic and non-phenolic compounds can be oxidized by lignin peroxidase that can cleave aryl Cα bond, Cα–Cβ bond, phenolic oxidation by aromatic ring opening, and finally demethylation.16 Lignin is a major contaminant in the textile, dye industry and is responsible for its intense unwanted dark-brown colour. Due to recalcitrant composition and corrosion resistance property, the non-hydrolysable bonds of lignin and some non-lignin dyes are resistant to degradation. The potential use of lignin-degrading bacteria and lignin peroxidase has become interesting because they can provide environment-friendly methods for dye-containing wastewater treatment of various industries. Here we will focus on some bacteria, fungi, or other microbes that can produce the lignin peroxidase enzyme and has dye decolourizing efficiency (Table).
Table :
Dye decolourization by lignin peroxidase producing microbes.
Microbial System |
Origin |
Decolourizing Dye |
Critical analysis of the Methods and Results |
References |
|---|---|---|---|---|
Kocuria rosea |
Gram-positive bacteria |
Azo dye, methyl orange |
Purified Lip decolourizes all dyes under long incubational conditions, but the heat-denatured enzyme did not decolourize tested dyes. |
[17] |
Phanerochaete chrysosporium |
Fungus |
Synthetic dyes |
The aga2 gene of P. chrysosporium has been inserted in the pCTCON2 and generated mutagenesis libraries, which decolourize diverse Azo dyes. |
[18] |
Nostoc musocorum, C. vulgaris |
Blue-green algae |
Naphthol Green B |
LiP reflects improved stability in partial purified enzymatic extraction (PPE) and degrades Malachite green, Naphthol green B, Brazillian wood, and orange G dyes. |
[16,19] |
Basidomycetes sp. |
Fungus |
Azo dye |
High efficiency of decolourization of the dye by Basidiomycetes sp. |
[20] |
P. Chrysosporium, B. adusta. |
Fungus |
Reactive violet |
Two Lip isoenzymes that have been purified here are similar to LiP isoenzymes from P. chrysosporium with respect to their catalytic properties and molecular mass. However, the B. adusta LiP isoenzymes are highly acidic than other fungi. |
[21] |
Genoderma sp., Phanerochaete |
Fungi |
Direct Violet, Reactive Black 5, Ponceau Xylidin, Diazo dyes, |
Azo dye degradation by different ligninolytic enzymes. |
[22] |
Alcaligenes aquatilis
|
Gram-negative bacteria |
Malachite green |
Alcaligenes aquatilisDB8 has been selected and identified by 16s rRNA, it has the capacity to decolourize different dyes including malachite green. |
[23] |
Funaliatrogii |
Fungi |
Methylene blue |
LiP has decolourized Methylene blue |
[24] |
Phanerochaete chrysosporium |
White rot fungi |
Crystal violet,
Methylene blue |
Crystal violet and Methylene blue degradation occur by LiP. |
[25] |
Phanerochaete sp. |
Crust fungi |
Melanin |
Lignin peroxidase enzyme effectively decolourizes melanin. |
[26] |
Daedoleopsis confragosa |
Fungi |
Navy blue HER, Orange HE2R |
D. confragosa is found to be new fungal isolates that have a role in dye decolourization. It has ligninolytic enzyme-like LiP suggests the possibility of commercialization of the production process. |
[27] |
Phanerochaete chrysosporium, N. fasciculare, F. rosea |
White rot fungi |
Congo red, Malachite green. |
Lignocellulosic waste contains different ligninolytic enzymes including LiP which plays a vital role in dye decolourization. |
[28] |
G. lucidum, F. fomentarius |
Fungi |
Congo Red, malachite green, and Methylene blue. |
Production of ligninolytic enzymes and breakdown of three aromatic dyes by using different white-rot fungi. |
[29] |
Pleurotus ostreatus PLO9, Ganoderma lucidum GRM117 |
Fungi |
RBBR dye |
LiP reflects improved stability in partial purified enzymatic extraction (PPE) and breaks RBBRdyes efficiently. |
[30] |
Manganese Peroxidase
Various unmodified microbial MnPs helps to breakdown approximately 60-99% dye by its decolourizing mechanism. Here are some examples that come from a different study. Microbial consortium SR can decolourise three different colours like Crystal Violet (63%) Cresol Red(93%), CBB G250(96%) within 6days with a minimum of 20 mg/L, 100 mg/L, 100 mg/L initial concentration of Dyes respectively.31 Trametes pubescens strain i8 can decolourise Acid Blue 158(95%), Poly R-478 (88%), Remazol Brilliant Violet 5R(76%), Direct Red 5B (66%), Indigo Carmine (64%), Methyl Green (50%), Cibacet Brilliant Blue BG (46%), Remazol Brilliant Blue (48%) with only 50 µM initial concentration in 24hours.32 Aspergillus terreus GS28 decolourise Direct Blue-1(98.4%) with 100 mg/L concentration within 168 hours.33 In the case of Bjerkandera adusta strain CX-9, Acid Blue 158(91%), Poly R-478(80%), Cibacet Brilliant Blue BG (77%), Remazol Brilliant Violet 5R (70%) at 50 µM initial concentration has been decolourised within 12 hours.34 Trametes sp. 48424 can decolourise 100 mg/L of Indigo Carmine(94.6%), Remazol Brilliant Blue R (85%), Remazol Brilliant Violet 5R (88.4%), Methyl Green (93.1%) within 18 hours.35 Microbial consortium ZSY can degrade the colour of the dye Metanil Yellow G(93.39%) at a minimum concentration of 100 mg/L within 48 hours.36 Microbial Consortium ZW1 decolourise the dye Methanil Yellow G(93.3%) with 100 mg/L in 16 hours37. Trichoderma harzianumcan break the colour of Blue-Black B(92.34%) at 0.03% initial concentration in 14 days.38 Phanerochaete chrysosporium CDBB 686 can decolourise with a 50-ppm concentration of Congo Red(41.84%), Poly R-478 (56.86%), Methyl Green (69.79%) within 36 hours39.Bjerkandera adusta CCBAS 930 can do the breakdown with a 0.01% concentration of Alizarin Blue Black B(86.5%), Acid Blue 129 (89.22%) by 20 days.40 white-rot fungus Cerrena unicolor BBP6 helps to break down six colours with 100 mg/L concentration. These are Congo Red (53.9% in 12 hours), Methyl Orange (77.6%in 12hours), Remazol Brilliant Blue R (81% in 5 hours), Bromophenol Blue (62.2% in 12 hours), Crystal Violet (80.9% in 12 hours), Azure Blue (63.1% within 24 hours).41 Phanerochaete chrysosporium breakdown 90.18% of Indigo Carmine with 30 mg/L initial concentration in 6 hrs.42 Ceriporia lacerata ZJSY decolourises 90% of Congo red with 100 mg/L concentration within 48 hrs.43 Bacillus cohnni RKS9 helps to break down 99% of Congo red with 100 mg/L concentrationin 12 hours.44 Schizophyllum commune IBL-06 decolourise 100% of Solar Brilliant Red 80 with 0.01% concentration within 3 days.45 Irpex lacteus CD2 decolourise different dye separately, Remazol Brilliant Violet 5R(92.8% in 5 hours), Remazol Brilliant Blue R (87.1% in 5 hours), Indigo Carmine (91.5% in 5 hours) Direct Red 5B (82.4% in 36 hours ) with 50mg/L initial concentration.46 On the other hand, Trametes versicolor can decolourise dye mixture 80.45% of Brilliant Blue FCF and Allura Red AC with an initial concentration of 100 mg/L within 14 days. Irpex lacteus can do 86.04% of dye decolourization in 19 days of this same dye mixture and Bjerkandera adusta do 82.83% dye degradation within 9 days.47
Cellulase
Cellulases are the complex group of enzymes that are secreted by a range of microorganisms which includes fungi, bacteria, and actinomycetes. In a natural environment, the interactions among the cellulolytic microorganisms result in the breakdown of lignocellulosic waste polymeric materials.48 Cellulase catalyzes the decomposition of cellulose by cleaving beta-1,4-glycosidic bonds. Complete hydrolysis of cellulose is mediated by the three enzymes. They are endoglucanase, exoglucanase, beta-glucosidase. The exoglucanase attacks the reducing end and non-reducing end of cellulose chains and produces glucose and cellobiose. The endoglucanase works against crystalline cellulose substrates such as cello-oligosaccharides, and beta-glucosidase hydrolyses cellobiose to glucose from the non-reducing ends.49 In fungi, the fungal Cellulases are secreted by Trichoderma reesei. In the case of actinomycetes, the genera produce cellulases are Streptomyces and Thermobifida, and for bacteria, it is Pseudomonas and Sphinomonas that produce cellulases. These are some important sources of enzymes that are used for textile dyes remediation. In the textile industry, cellulases are used as dye decolourization enzymes. Currently, in the textile industry cellulases are best applicable in the bio-stoning and biopolishing process.50 Microbial cellulases are an alternative to the traditional method of bio-stoning. Cellulases act on cotton of denim fabric. The indigo dye is used for the colouration of the denim fabric. The dye is trapped inside the cellulose fibre in the cotton material. The dye is mostly attached to the surface of the yarn and most exterior of short cotton fibres. Cellulases hydrolyze and breaks the small fibres of fabric by breaking the beta-1,4- linkages of the cellulose. This hydrolysis process removes the fibres which trap indigo dye. The dye is easily removed from the fabric after this process.48,51 Trichoderma reesei endoglucanase II is the best suitable candidate for bio-stoning. Cellulases have several advantages and disadvantages over the conventional approach which is a stonewash with a pumice stone. The advantages of using cellulases are: it gives high productivity, less work intensiveness, are safer for the environment, it takes short time than the conventional method. There are several disadvantages also. The major disadvantage of cellulase is back staining which is the redeposition of dye on the fabric and losses of the shade look given by the treatment.51 The latest trend of bio stone washing is using an enzyme mixture composed of amylase, cellulases, lactase.52 Cellulases also play a critical role in biopolishing where they remove excess stain from the denim fabric. Apart from indigo dye, cellulases are involved in various dye decolourizations (Table 2). Dye like methylene blue, malachite green, Congo red, methyl orange dye, grams iodine dye is decolourized by Cellulase enzymes under optimum conditions like temperature, pH, and time.53
Table (2):
Dye decolourization by cellulase enzyme.
Dyes |
The optimum temperature for dye decolourization |
Time taken for dye decolourization |
Optimum pH for dye decolourization |
Cellulases Producing Bacteria/ Fungi |
References |
|---|---|---|---|---|---|
Congo red |
26°C |
2 – 5 days |
pH 4.5 |
Trichoderma reesei |
[54] |
Malachite green |
40°C |
1200 min |
pH 11 |
Bacillus cereus |
[53] |
Grams Iodine |
28°C |
30 – 40 min |
pH 7.0 |
Bacillus sp, Pseudomonas sp |
[55] |
Drimarene dye |
28°C |
25 min |
pH 2.0 |
Trichoderma reesei, Streptomyces and Thermobifida |
[56] |
Pectinase
Pectinase enzymes act on pectin by cleaving the glycosidic bond of galacturonic acid.57 Pectins are the chain molecules with a rhamnogalacturonan backbone associated with other polymers and carbohydrates.58 Pectins are heteropolysaccharide structures made up of alpha (1,4) linked D-galacturonic acid residues.59 Pectinase enzymes depolymerize pectin through hydrolysis, trans-elimination, and de-esterification mechanisms. These reactions hydrolyze the ester bond of pectin.58 Pectinase enzymes are classified according to their mode of action like 1: Methylesterases, remove methoxy groups from esterified galacturonan. 2: Polygalacturonases, that is subdivided into endopolygalacturonase (catalyzes the hydrolysis of the glycosidic bond randomly), and exopolygalacturonase, which releases galacturonic acid residues from the non-reducing ends of homogalacturonan.59 Pectinase has an important role in the food industry and is commercially used for juice extraction, wine clarification, and decolourization of both.60 The microbial sources of pectinase enzymes are Aspergillus niger, Aspergillus oryzae, Penicillium restrictum, Trichoderma viridae, Bacillus subtilis, and Bacillus cereus.61 Pectins and various polysaccharides are the substances present in fruit, they lead to colloid formation and fouling, also reduce the commercial value of juices.62 Pectinase degrades the pectin by cleaving beta-1,4-glycosidic bonds present in pectin. It reduces viscosity and cluster formation in juices that enhance the clarity of juices.63 The decolourization process is important to give a pleasant colour of fruit juices.64 Pectinase enzyme helps in the decolourization of Congo red dye. This decolourization occurs under certain conditions like at pH 6 – 6.5, temperature 28°C, and it takes 5 days to decolourize the congored by the microorganism Aspergillus oryzae by forming a zone on congo red-agar medium.65
Dye Decolourization By Microbial Consortia
Microbial consortia meaning is when two or more microbial groups live together symbiotically. Microbial consortia can be ectosymbiotic or endosymbiotic or sometimes maybe both. The evolution of land plants and their transition from algal communities in the sea to land microbial consortia suggest symbiotic evidence between their necessary precursors. Microbial consortia can decolourize dyes (Table 3); mostly the synthetic dye(azo dyes) that are present in the industrial effluents. Azo dyes contain one or more –N=N- groups which are commonly found in synthetic group release in nature. Azo bond is metabolized by reductive cleavage while the consequent aromatic amines are metabolized under gaseous conditions. Hence, the microbial population of the treatment system should work under both anaerobic/ anoxic and gaseous conditions to gain complete mineralization of dye molecules. By using microbial consortia, the azo dye decolourization occurs faster. Dye contaminated soil has been isolated from textile wastewater of Orissa, India, that contains the pure culture of bacterial consortium-BP of Bacillus flexus TS8(BF), Proteus mirabilis PMS(PM), and Pseudomonas aeruginosa NCH (PA). Physico-chemical parameters have been optimized to gain maximum discolouration efficiency. The formation of metabolites by degradation of Indanthrene Blue RS has been confirmed through UV-Vis spectroscopy, FT-IR, and GC-MS analysis. When the agricultural residual wastes have been supplemented, it shows an enhanced decolourization efficiency of consortium-BP. Mineralization of Indanthrene Blue RS has determined the higher reduction in TOC(Total Organic Carbon). COD(Chemical Oxygen Demand) has been removed by consortium-BP. Bacillus flexus, Proteus mirabilis, and Pseudomonas aeruginosa show a positive result in the catalase test. Bacterial consortia of Pseudomonas aeruginosa, Rhodobacter sphaeroides, Proteus mirabilis, Bacillus circulans have the reaction in anoxic-oxic condition to decolourize Renazol Black B. They incubate the consortia to observe the colour reduction, and they found that 90% of colour reduction and a COD reduction of 80% occur by using synthetic wastewater with a dye concentration of 100mgL-1. For decolourization of golden yellow HER dye under aerobic and microaerophilic conditions, Microbial consortia GG-BL consisting of Galactomyces geotricium and Brevibacillus laterosporus NCIM 2298 has been developed. They have catalase, reductase enzymes. Bacterial consortia Zobellelata iwanensis ATI-3 and Bacillus pumilus HKG212 are used under static conditions to decolourize Reactive green -19 dyes. Yeast extract has been added as co-substrate, the decolourization efficiency of 97% with initial dye concentration has been observed. It is difficult to determine the impact of the experimental condition and decolourization process together.66 Textile Acid Orange dye from textile effluent contaminated soil of Tanda, Uttar Pradesh(India) has been isolated. This dye is decolourized by a bacterial strain RMLRT03. Bushnell and Haas medium (BHM) amended with Acid Orange dye has been used for decolourization studies. 16s rRNA sequence of the bacterial strain identifies it as Staphylococcus hominis.67 This bacterial strain has good decolourization ability with glucose and yeast extract supplements as co-substrate in static conditions. The optimal conditions of Acid Orange dye decolourization are at pH 7.0 and 35°C in 60 hours incubation by Staphylococcus hominis. The textile dyes can be absorbed or degraded by many bacterial and fungal species. Anthraquinone dye can be decolourized either by aerobic or anaerobic conditions. They are resistant to corrosion due to their mixed aromatic structure. It lasts for a long time and causes considerable concern for environmental pollution and waste retention. Bioremediation techniques have attracted a lot of attention, making microbial discolouration and degradation economical and environmentally friendly compared to various conventional methods. Some potential bacteria can fade synthetic/ commercial dyes are used in textile dying. Here firstly five bacterial consortia have been isolated (Bacillus sp.1, Bacillus sp.2, Acinetobacter sp., Citrobacter sp., and Klebsiella sp.). Now they are divided into three groups; (a) Bacillus sp.1, Bacillus sp.2, Acinetobacter sp., Citrobacter sp. (b) Acinetobacter sp., Citrobacter sp., Bacillus sp1., (c) five bacterial consortia. The results show that a mixture of five bacterial consortia has more efficiency to decolourize the dye.68 Bacillus cohnii, Aspergillus terreus HTCC, Penicillumcitrinum are able to decolourize Basic violet dye. Various parameters like initial dye density, dye to inoculum ratio, and incubation time duration has been studied for dye discolouration. The evolving fungal bacterial association exhibits the highest percentage of discolouration (92%) ability compared to dye treatment by the monoculture approach. Fungal –Bacterial (Penicillumcitrinum and Bacillus cohnii) consortia are more efficiently decolourized Basic violet dye. An integrated degradation and detoxification of textile dyes may be possible by the combination of fungi and bacteria that provide a good alternative technology for contaminant removal of water. To degrade the textile effluent dyes basically the Acid dyes by bacterial and fungal consortia, these isolates have been used to form a mixed microbial consortium cell factory that could quickly fade and biodegrade the organic load on the waste material and be used to develop a continuous process for the treatment of a variety of textile dyed textile wastes, including reactive dyes.
Table (3):
Dye decolourization by Microbial Consortia.
Dye |
Microbial Source |
Microbes Name |
Consortia |
Decolorizing (%) |
Time |
Enzyme |
Reference |
|---|---|---|---|---|---|---|---|
Acid Red 88 |
Local textile industry effluent in Punjab, India |
Bacillus cereus (BN-7), Pseudomonas fluorescence (BN-5), Stenotrophomonas acidaminiphila (BN-3) |
Bacterial consortia |
78-94% |
24 hrs |
Azo-reductase |
[69] |
Mono azo,
DI-azo, tri-azo |
Textile effluent |
Klebsiella sp.1, Klebsiella sp.2, Staphylococcus aureus, Bacillus cereus, Pseudomonas fluorescence |
Bacterial consortia |
Yellow dye 63%, green dye 92%, orange dye 40%, blue dye 65%, black dye 96% |
15 days |
catalase |
[70] |
Azo dyes (DB151 and DR31) |
Textile effluent |
Bacteroides spp. ,Eubacterium spp., Clostridium spp., Proteus vulgaris , Streptococcus faccalis |
Bacterial consortia |
95.25%-98% |
5 days |
Catalase, oxidase |
[71,72] |
Azo dyes |
Industrial effluent |
Acinetobacter sp., Klebsiella sp., Pseudomonas sp. |
Bacterial consortia |
80% |
72hrs |
Peroxidase |
[71,73] |
Reactive Green -19 |
Textile wastewater |
Zobellelata iwanensis,, Bacillus pumilus |
Bacterial consortia |
86-93% |
24 hrs |
laccase |
[71,74] |
Disperse Red |
Textile effluent |
Microbacterium sp.,
Leucobacter albus, Klebsiella sp., Staphylococcus arlettae |
Bacterial consortia |
80% |
72 hrs |
Azo reductase |
[71,75] |
Reactive black 5 |
Textile effluent |
Anaxybacillus flavithermus, Anaxybacilluskamehatkensis, Anaxybacilluspushchinoensis |
Bacterial consortia |
70% |
24hrs |
Azo reductase |
[76] |
RB-5 |
Textile wastewater |
Pseudoarthrobacter, Gordonia, Stenotrophomonas, Sphingomonas |
Bacterial consortia |
54-34% |
7 days |
Azo reductase |
[76] |
Azo blue and Red dye |
Polluted water |
Enterobacter aerogens, Proteus rettgeri, Pseudomonas fluorescens |
Bacterial consortia |
80-90% |
96 hrs |
Azo reductase |
[77] |
Red, green, yellow, black Azo dyes |
Effluent from Hudiara drain near Nishat mills ltd, Lahore |
Bacillus subtilis ,Bacillus areus, Bacillus mycoides, Bacillus sp.,Micrococcus sp., Pseudomonas sp. |
Bacterial consortia |
84% |
24
hrs |
reductase |
[68] |
Safranin, congo red and CBBG-250 |
Municipal sewage system waste water |
Bacillus subtilis, Aeromonas hydrophila, Proteus mirabilis, Pseudomonas sp., Shewanella sp. |
Bacterial consortia |
90% |
36
hrs |
reductase |
[78] |
Azo dye |
Textile effluent Bangladesh |
Bacillus thuringiensis, Bacillus pumilus , Enterococcus faccium |
Bacterial consortia |
77.1% |
72
hrs |
Azo reductase |
[79] |
Reductive red dye |
Textile effluent |
Bacillus subtilis, A. niger |
Bacterial-fungal consortium |
90% |
72
hrs |
Laccase, azo reductase, NADH-DCIP reductase |
[80] |
Orange, G-red |
Textile effluent |
Chlorella vulgaris, Lyngbya sp., Nostoc lincui, Elnatothrixviridis, Volvox sp., |
Bacterial-algal consortium |
47-59% |
7 days |
Laccase, peroxidase |
[81] |
Methyl red |
Textile effluent |
Klebsiella sp., Bacillus sp., Clostridium sp., |
Bacterial consortium |
100% |
16hrs |
Azo reductase |
[82] |
Yellow EXF, Red EXF |
Textile effluent |
Proteus mirabilis, Morganella morganii and Enterobacter cloacae |
Bacterial consortium |
94% |
72hrs |
Laccase, azo reductase, oxidase |
[83] |
Acid blue |
Textile effluent |
Citrobacter freundii, Moraxella osloensis, Pseudomonas aeruginosa, Citrobacter sp., |
Bacterial consortium |
97% |
48hrs |
Azo reductase |
[84] |
Azo dyes |
Wastewater |
Bacillus subtilis, Achromobacterxyloxidans |
Bacterial consortia |
70% |
7 days |
Laccase |
[85] |
Direct Red 28 |
Wastewater |
Bacillus pumilus., B. cereus, B.subtilis and B. megaterium. |
Bacterial consortia |
72-92% |
10 days |
Azo reductase |
[86] |
Reactive blue 160 |
Textile effluent |
Alcaligenes sp., Bacillus sp. BAB2731, Escherichia sp. BAB2734, Pseudomonas sp. BAB3054, Providencia sp. BAB2749, Acinetobacter sp. BAB2750, Bacillus sp. BAB2751, Bacillus sp. BAB3055 |
Bacterial consortium |
100% |
4hrs |
Azo reductase, lignin peroxidase |
[87] |
Acid Red 88 |
Local textile industry effluent in Punjab, India |
Bacillus cereus (BN-7), Pseudomonas fluorescence (BN-5), Stenotrophomonas acidaminiphila (BN-3) |
Bacterial consortia |
78-94% |
24 hrs |
Azo-reductase |
[69] |
Mono azo, DI-azo, tri-azo |
Textile effluent |
Klebsiella sp.1, Klebsiella sp.2, Staphylococcus aureus, Bacillus cereus, Pseudomonas fluorescence |
Bacterial consortia |
Yellow dye 63%, green dye 92%, orange dye 40%, blue dye 65%, black dye 96% |
15 days |
catalase |
[70] |
Azo dyes (DB151 and DR31) |
Textile effluent |
Bacteroides sp., Eubacterium spp., Clostridium spp., Proteus vulgaris, Streptococcus faccalis |
Bacterial consortia |
95.25%-98% |
5 days |
Catalase, oxidase |
[71, 72] |
Azo dyes |
Industrial effluent |
Acinetobacter sp., Klebsiella sp., Pseudomonas sp |
Bacterial consortia |
80% |
72hrs |
Peroxidase |
[71,73] |
Reactive Green -19 |
Textile wastewater |
Zobellelata iwanensis,, Bacillus pumilus |
Bacterial consortia |
86-93% |
24 hrs |
laccase |
[71,74] |
Genetically Engineered Microorganisms And Their Impact On Dye Decolourization
Dye is a natural or chemical ingredient that imparts colour when applied to something. It is of two types, natural and synthetic. Synthetic dyes are broadly used in different industries like textile, paper, leather, food, pharmaceutical industries. These dyes have replaced natural dyes over the past few years due to their wide variety of colours, low cost, and capacity to withstand damage by sunlight, water, and chemicals.88 Dyes are categorized into 14 types based on their structure. These are Acid dyes, direct dyes, and azo dyes, disperse dyes, sulfur dyes, fibre reactive dyes, basic dyes, oxidation dyes, mordant dyes, developed dyes, vat dyes, pigments, fluorescence or optical brighteners, and solvent dyes. Azo dyes are the largest group of synthetic dyes with more than 2000 different types. These are substantially used in the textile industry due to their bright colour, water fastness, and simple application technique. Although dyes are greatly used in industries, the intense usage of synthetic dyes has augmented water pollution. Dyes have a great solubilizing capability in water, which makes it so difficult to be removed from water.89 According to WHO, dyeing treatment in the textile industry causes 17-20% of the industrial water pollution and among these dyes, 80% is azo dyes.90 Synthetic nitrogen-based dyes are so toxic that they are banned in European Union, China, Japan, India, and Vietnam. The toxic effect of the dyes causes damage to the flora and fauna including humans. Therefore, the degradation and decolourization of these dyes are very important. The traditional dye decolourization technology involves physical (flocculation, coagulation, adsorption etc.), chemical (precipitation, oxidation), and biological (microbes, enzymes, microbial fuel cells etc.) methods.91 Biological methods involve the use of microorganisms and their pathways to perform the decolourization of dyes. Averse to the physical and chemical methods, biological methods are more efficient, eco-friendly, and versatile. Biological methods using microorganisms are advantageous over the others because it is inexpensive, low cost and completely mineralize the organic pollutants. Microorganisms like bacteria, fungi, yeast, algae possess the ability to decolourize dyes. Genetic engineering plays a significant role in dye decolourization (Table 4). The recent advancement in molecular biology and genetic engineering has opened a new way to fight the pollution problem caused by these dyes. Each microorganism has a different capability for dye degradation and bioremediation.92 GMOs can be made by transferring a specific gene from one species to another or by gene modification. Genetically engineered microorganisms possess enhanced dye decolourization or bioremediation capacity. Insertion of various naturally occurring genes in a suitable host linked to several enzymatic activities resulting in the expression of designed pathways leading to the degradation of these dyes could be a useful tool for reducing pollution.
Table (4):
Genetically engineered microorganisms and their impact on dye decolourization.
| Gene name | Extracted from | Expressed in | Vector | Dye | Reference |
|---|---|---|---|---|---|
| azoreductase | Rhodobacter sphaeroides AS 1.1737 | Escherichia coli JM109 | pGEX4T-1 | Acid red GR | [93] |
| azoreductase | Rhodobacter sphaeroides AS 1.1737 | Escherichia coli JM109 | pGEX4T-1 | C.I. Direct blue 71 | [94] |
| azoreductase | Rhodobacter sphaeroides AS1.1737 | Escherichia coli JM109 | pGEX4T-1 | Acid red B | [95] |
| azoreductase | Halomonas elongata | Escherichia coli DH5 | pET21a | Methyl red and Remazol Black B | [96] |
| azrS | Bacillus sp. MR-1/2 | Escherichia coli DH5a | – | Congo red And reactive black 5 | [97] |
| sodC | Synechococcus sp. PCC 9311 | Synechococcus elongatus PCC 7942 | pSyn_6 | Acid black 1 | [98] |
| azoreductase | Bacillus latrosporus RRK1 | Escherichia coli DH5a | pAZR-SS125 | Remazol red | [99] |
| laccase | Bacillus subtilis CotA | Synechococcus elongatus PCC 7942 | pCV0062 | Indigo carmine, Reactive blue 19, Reactive black 5, Reactive blue 4 | [100] |
| add | Rhodococcus sp. | Escherichia coli DH5a | pAZRS1 | Reactive red 22 | [101] |
| azoG | Halomonas sp. Strain GT | Escherichia coli DH5a | pET30a(+) | Azo dye wastewater | [102] |
| cotA | Bacillus subtilis 168 | Escherichia coli | pET-28a(+) | Malachite green, Acid blue 62 and Methyl orange | [103] |
| – | Pseudomonas sp. SUK1 | Escherichia coli | – | Red BLI, Navy blue-HER and Golden yellow-HER | [104] |
| azoA | Enterococcus sp. L2 | Escherichia coli | pBBR1MCS2fdh-azoA | Reactive red 97, acid red 119, reactive black B5, like azo dyes | [105] |
| Pseudomonas fluorescens | pBBR1MCS2 azoA |
Enzymes are essential in many biological processes, the role of the lignocellulolytic enzyme in dye decolourization is very important. Dye decolourization is necessary because the dye is toxic in nature. The removal of dye contaminants from waste effluents using microorganism-derived lignocellulolytic enzymes has shown promising results with maximum efficiency because they show outstanding decolourization capabilities for various classes of dyes and could be used in place of synthetic dye decolourizing agents. Decolourization of the dyes will reduce their biological load. In the future, scientists can develop a genetically modified consortium that will contain all the lignocellulolytic enzymes which will help in dye decolourization. This will make the job easier and less time-consuming. Bio-decolourization has gained importance as an alternative, eco-friendly, low-cost, and efficient technology for industrial dye removal treatment. In addition, genetic, metabolic engineering technologies, Omics strategies, and synthetic biology approaches have significantly enhanced the stability and capacity of biocatalysts that reduce the reaction time. Still, many challenges are limiting large-scale commercial production. Future research should be carried out to determine (1) dye-degradation mechanisms, process parameter optimization for microbial growth; (2) Response Surface Methodology (RSM) generation for consortia parameters to reduce enzyme loss and improve enzymes durability; (3) improve catalytic domain for enhancement of catalytic performance by in-silico protein-ligand interaction model generation in the dry laboratory and to achieve this, mutagenesis study in the wet laboratory.
ACKNOWLEDGMENTS
The authors would like to thank JIS University Kolkata and JIS Group Educational Initiatives.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
DG, SD conceptualized the study. SD, AG, AC, SM, SR wrote the manuscript. DG reviewed and edited the manuscript. All authors read and approved the final manuscript for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- Chukwuma OB, Rafatullah M, Tajarudin HA, Ismail N. Lignocellulolytic enzymes in biotechnological and industrial processes: A review. Sustainability. 2020;12(18):7282.
Crossref - Affat SS. Classifications, advantages, disadvantages, toxicity effects of natural and synthetic dyes: A review. Edu.iq. https://www.jsci.utq.edu.iq/index.php/main/article/download/790/733. Accessed May 30, 2022.
- Chavan RB. Health and environmental hazards of synthetic dyes. Textile review magazine, May. Published online 2013:12-17. https://static.fibre2fashion.com/ArticleResources/PdfFiles/70/6909.pdf
- Singh RS, Singh T, Pandey A. Microbial Enzymes-An Overview. Advances in Enzyme Technology. 2019:1-40.
Crossref - Nunes CS, Kunamneni A. Laccases-properties and applications. Enzymes in Human and Animal Nutrition. 2018:133-161.
Crossref - Asadi E, Makhdoumi A, Asoodeh A. Laccase mediator system obtained from a marine spore exhibits decolorization potential in harsh environmental conditions. Ecotoxicol Environ Saf. 2020;191:110184.
Crossref - Singh G, Bhalla A, Kaur P, Capalash N, Sharma P. Laccase from prokaryotes: a new source for an old enzyme. Rev Environ Sci Biotechnol. 2011;10(4):309-326.
Crossref - Koschorreck K, Schmid RD, Urlacher VB. Improving the functional expression of a Bacillus licheniformislaccase by random and site-directed mutagenesis. BMC Biotechnol. 2009;9(1):12.
Crossref - Wang CL, Zhao M, Li DB, Cui DZ, Lu L, Wei XD. Isolation and characterization of a novel Bacillus subtilis WD23 exhibiting laccase activity from forest soil. Ajol.info.2010;9(34):5496-5502 https://www.ajol.info/index.php/ajb/article/view/92100/81539. Accessed May 30, 2022.
- Hullo MF, Moszer I, Danchin A, Martin-Verstraete I. CotA of Bacillus subtilis is a copper-dependent laccase. J Bacteriol. 2001;183(18):5426-5430.
Crossref - Chen S, Ge W, Buswell JA. Molecular cloning of a new laccase from the edible straw mushroom Volvariella volvacea: possible involvement in fruit body development. FEMS Microbiol Lett. 2004;230(2):171-176.
Crossref - Lu L, Zhao M, Wang TN, et al. Characterization and dye decolorization ability of an alkaline resistant and organic solvents tolerant laccase from Bacillus licheniformis LS04. Bioresour Technol. 2012;115:35-40.
Crossref - Reiss R, Ihssen J, Thony-Meyer L. Bacillus pumilus laccase: a heat stable enzyme with a wide substrate spectrum. BMC Biotechnol. 2011;11(1):9.
Crossref - Blanquez A, Rodriguez J, Brissos V, et al. Decolorization and detoxification of textile dyes using a versatile Streptomyces laccase-natural mediator system. Saudi J Biol Sci. 2019;26(5):913-920.
Crossref - Dube E, Shareck F, Hurtubise Y, Daneault C, Beauregard M. Homologous cloning, expression, and characterisation of a laccase from Streptomyces coelicolor and enzymatic decolourisation of an indigo dye. Appl Microbiol Biotechnol. 2008;79(4):597-603.
Crossref - Abd Ellatif S, El-Sheekh MM, Senousy HH. Role of microalgal ligninolytic enzymes in industrial dye decolorization. Int J Phytoremediation. 2021;23(1):41-52.
Crossref - Parshetti GK, Parshetti S, Kalyani DC, Doong RA, Govindwar SP. Industrial dye decolorizing lignin peroxidase from Kocuria rosea MTCC 1532. Ann Microbiol. 2012;62(1):217-223.
Crossref - Durdic KI, Ostafe R, Prodanovic O, et al. Improved degradation of azo dyes by lignin peroxidase following mutagenesis at two sites near the catalytic pocket and the application of peroxidase-coated yeast cell walls. Front Environ Sci Eng. 2021;15(2):19.
Crossref - Chantarasiri A, Boontanom P. Decolorization of synthetic dyes by ligninolytic Lysinibacillus sphaericus JD1103 isolated from Thai wetland ecosystems. Aquaculture, Aquarium, Conservation & Legislation. 2017;10(4):814-819.
- Eichlerova I, Baldrian P. Ligninolytic enzyme production and decolorization capacity of synthetic dyes by saprotrophic white rot, brown rot, and litter decomposing Basidiomycetes. J Fungi. 2020;6(4):301.
Crossref - Heinfling A, Martinez MJ, Martinez AT, Bergbauer M, Szewzyk U. Purification and characterization of peroxidases from the dye-decolorizing fungus Bjerkandera adusta. FEMS Microbiol Lett. 1998;165(1):43-50.
Crossref - Chaturvedi S. Azo dyes decolorization using white rot fungi. Research & Reviews: J Microbiol Biotechnol. 2019;8(2):9-19. https://www.rroij.com/open-access/azo-dyes-decolorization-using-white-rot-fungi.php?aid=87649. Accessed May 30, 2022.
- Mh Rizk N, Eldourghamy A, Aly S, Sabae S, Sobhy A. Production of lignin peroxidase from aquatic bacteria, Alcaligenes aquatilis. Egyptian Journal of Aquatic Biology and Fisheries. 2020;24(3):213-223.
Crossref - da Silva MR, de Sa LRV, Russo C, Scio E, Ferreira-Leitao VS. The use of HRP in decolorization of reactive dyes and toxicological evaluation of their products. Enzyme Res.;2010:703824.
Crossref - Mary JE, Krithika T, Kavitha R. Biodegradation of textile dye by ligninolytic bacteria isolated from Western Ghats. Int J Res Rev. 2020;7(4):22-29.
- Sung HJ, Khan MF, Kim YH. Recombinant lignin peroxidase-catalyzed decolorization of melanin using in-situ generated H2O2 for application in whitening cosmetics. Int J Biol Macromol. 2019;136:20-26.
Crossref - Manawadi SI, Vantamuri AB, Guruvin SK. Characterization of ligninolytic enzymes and decolourization of selected textile dyes from the blushing bracket mushroom, Daedaleopsis confragosa. Int J Pharm Sci & Res. 2019;10(12):5592-5598.
Crossref - Kumar A, Chandra R. Ligninolytic enzymes and its mechanisms for degradation of lignocellulosic waste in environment. Heliyon. 2020;6(2):e03170.
Crossref - Jayasinghe C, Imtiaj A, Lee GW, et al. Degradation of three aromatic dyes by white rot fungi and the production of ligninolytic enzymes. Mycobiology. 2008;36(2):114-120.
Crossref - Oliveira SF, da Luz JMR, Kasuya MCM, Ladeira LO, Correa Junior A. Enzymatic extract containing lignin peroxidase immobilized on carbon nanotubes: Potential biocatalyst in dye decolourization. Saudi J Biol Sci. 2018;25(4):651-659.
Crossref - Yang X, Wang J, Zhao X, Wang Q, Xue R. Increasing manganese peroxidase production and biodecolorization of triphenylmethane dyes by novel fungal consortium. Bioresour Technol. 2011;102(22):10535-10541.
Crossref - Rekik H, Zarai Jaouadi N, Bouacem K, et al. Physical and enzymatic properties of a new manganese peroxidase from the white-rot fungus Trametes pubescens strain i8 for lignin biodegradation and textile-dyes biodecolorization. Int J Biol Macromol. 2019;125:514-525.
Crossref - Singh G, Dwivedi SK. Decolorization and degradation of Direct Blue-1 (Azo dye) by newly isolated fungus Aspergillus terreus GS28, from sludge of carpet industry. Environ Technol Innov. 2020;18:100751.
Crossref - Bouacem K, Rekik H, Jaouadi NZ, et al. Purification and characterization of two novel peroxidases from the dye-decolorizing fungus Bjerkandera adusta strain CX-9. Int J Biol Macromol. 2018;106:636-646.
Crossref - Zhang H, Zhang S, He F, Qin X, Zhang X, Yang Y. Characterization of a manganese peroxidase from white-rot fungus Trametes sp.48424 with strong ability of degrading different types of dyes and polycyclic aromatic hydrocarbons. J Hazard Mater. 2016;320:265-277.
Crossref - Guo G, Hao J, Tian F, et al. Decolorization and detoxification of azo dye by halo-alkaliphilic bacterial consortium: Systematic investigations of performance, pathway and metagenome. Ecotoxicol Environ Saf. 2020;204:111073.
Crossref - Guo G, Hao J, Tian F, et al. Decolorization of Metanil Yellow G by a halophilic alkali thermophilic bacterial consortium. Bioresour Technol. 2020;316:123923.
Crossref - Rybczynska-Tkaczyk K, Kornillowicz-Kowalska T, Szychowski KA, Gminski J. Biotransformation and toxicity effect of monoanthraquinone dyes during Bjerkandera adusta CCBAS 930 cultures. Ecotoxicol Environ Saf. 2020;191:110203.
Crossref - Sosa-Martinez JD, Balagurusamy N, Montanez J, et al. Synthetic dyes biodegradation by fungal ligninolytic enzymes: Process optimization, metabolites evaluation and toxicity assessment. J Hazard Mater. 2020;400:123254.
Crossref - Rybczynska-Tkaczyk K, Swiecilo A, Szychowski KA, Kornillowicz-Kowalska T. Comparative study of eco- and cytotoxicity during biotransformation of anthraquinone dye Alizarin Blue Black B in optimized cultures of microscopic fungi. Ecotoxicol Environ Saf. 2018;147:776-787.
Crossref - Zhang H, Zhang J, Zhang X, Geng A. Purification and characterization of a novel manganese peroxidase from white-rot fungus Cerrena unicolor BBP6 and its application in dye decolorization and denim bleaching. Process Biochem. 2018;66:222-229.
Crossref - Li H, Zhang R, Tang L, Zhang J, Mao Z. Manganese peroxidase production from cassava residue by Phanerochaete chrysosporium in solid state fermentation and its decolorization of indigo carmine. Chin J Chem Eng. 2015;23(1):227-233.
Crossref - Wang N, Chu Y, Wu F, Zhao Z, Xu X. Decolorization and degradation of Congo red by a newly isolated white rot fungus, Ceriporia lacerata, from decayed mulberry branches. Int Biodeterior Biodegradation. 2017;117:236-244.
Crossref - Kishor R, Purchase D, Saratale GD, et al. Environment friendly degradation and detoxification of Congo red dye and textile industry wastewater by a newly isolated Bacillus cohnni (RKS9). Environ Technol Innov. 2021;22:101425.
Crossref - Asgher M, Yasmeen Q, Iqbal HMN. Enhanced decolorization of Solar brilliant red 80 textile dye by an indigenous white rot fungus Schizophyllum commune IBL-06. Saudi J Biol Sci. 2013;20(4):347-352.
Crossref - Qin X, Zhang J, Zhang X, Yang Y. Induction, purification and characterization of a novel manganese peroxidase from Irpex lacteus CD2 and its application in the decolorization of different types of dye. PLoS One. 2014;9(11):e113282.
Crossref - Merino-Restrepo A, Mejia-Otalvaro F, Velasquez-Quintero C, Hormaza-Anaguano A. Evaluation of several white-rot fungi for the decolorization of a binary mixture of anionic dyes and characterization of the residual biomass as potential organic soil amendment. J Environ Manage. 2020;254:109805.
Crossref - Jayasekara S, Ratnayake R. Microbial Cellulases: An Overview and Applications. In: Cellulose. IntechOpen; 2019.
Crossref - Gupta P, Samant K, Sahu A. Isolation of cellulose-degrading bacteria and determination of their cellulolytic potential. Int J Microbiol. 2012;1-5.
Crossref - Kuhad RC, Gupta R, Singh A. Microbial cellulases and their industrial applications. Enzyme Res. 2011;280696.
Crossref - Behera BC, Sethi BK, Mishra RR, Dutta SK, Thatoi HN. Microbial cellulases – Diversity & biotechnology with reference to mangrove environment: A review. J Genet Eng Biotechnol. 2017;15(1):197-210.
Crossref - Maryan AS, Montazer M. A cleaner production of denim garment using one step treatment with amylase/cellulase/laccase. J Clean Prod. 2013;57:320-326.
Crossref - Wanyonyi WC, Onyari JM, Shiundu PM, Mulaa FJ. Enzymatic decolorization of malachite green dye by a newly isolated Bacillus cereus strain wwcp1. IOSR J Environ Sci Toxicol Food Technol (IOSR-JESTFT). 2014;2(12):58-64.
Crossref - Sazci A, Erenler K, Radford A. Detection of cellulolytic fungi by using Congo red as an indicator: a comparative study with the dinitrosalicyclic acid reagent method. J Appl Bacteriol. 1986;61(6):559-562.
Crossref - Kasana RC, Salwan R, Dhar H, Dutt S, Gulati A. A rapid and easy method for the detection of microbial cellulases on agar plates using gram’s iodine. Curr Microbiol. 2008;57(5):503-507.
Crossref - Vyjayanthi JP, Suresh S. Decolorization of drimarene red dye using palladized bacterial cellulose in a reactor. Water Environ Res. 2010;82(7):601-609.
Crossref - Ubani CS, Ezugwu AL, Oje OA, Gabriel SC, Ekwedigwe AM. Isolation, partial purification and characterization of pectinase from water melon (Citrullus lanatus) rind. American-Eurasian Journal of Toxicological Sciences. 2015;7(2):110-114.
Crossref - Saranraj P, Naidu MA. Microbial pectinases: a review. Global J Trad Med Syst. 2014;3(1):1-9. https://www.researchgate.net/publication/261098549_Microbial_Pectinases_A_Review.
- Kc S, Upadhyaya J, Joshi DR, et al. Production, characterization, and industrial application of pectinase enzyme isolated from fungal strains. Fermentation. 2020;6(2):59.
Crossref - Kubra KT, Ali S, Walait M, Sundus H. Potential applications of pectinases in food, agricultural and environmental sectors. J Pharm Chem Biol Sci. 2018;6(2):23-34. https://www.jpcbs.info/2018_6_2_03_%20Kubra.pdf.
- Bhardwaj V, Degrassi G, Bhardwaj RK. Microbial pectinases and their applications in industries: a review. Polymer. 2017;4(08). https://www.irjet.net/archives/V4/i8/IRJET-V4I8144.pdf.
- Nawaz A, Sameer M, Akram F, et al. Kinetic and thermodynamic insight of a polygalacturonase: A biocatalyst for industrial fruit juice clarification. Rev Mex Ing Quim. 2021;20(2):1029-1045.
Crossref - Anand G, Yadav S, Yadav D. Purification and characterization of polygalacturonase from Aspergillus fumigatus MTCC 2584 and elucidating its application in retting of Crotalaria juncea fiber. 3 Biotech. 2016;6(2):201.
Crossref - Dey TB, Banerjee R. Application of decolourized and partially purified polygalacturonase and ב-amylase in apple juice clarification. Braz J Microbiol. 2014;45(1):97-104.
Crossref - Akbar S, Prasuna RG. Exploitation of fruit wastes for pectinase production using Aspergillus oryzae. Int J Pharm Bio Sci. 2012;3(3):756-765. https://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.640.4946&rep=rep1&type=pdf.
- Mohanty SS, Kumar A. Enhanced degradation of anthraquinone dyes by microbial monoculture and developed consortium through the production of specific enzymes. Sci Rep. 2021;11(1):7678.
Crossref - Singh RP, Singh PK, Singh RL. Bacterial decolorization of textile azo dye Acid Orange by Staphylococcus hominis RMLRT03. Toxicol Int. 2014;21(2):160-166.
Crossref - Mahmood R, Sharif F, Ali S, Hayyat MU. Enhancing the decolorizing and degradation ability of bacterial consortium isolated from textile effluent affected area and its application on seed germination. Scientific World Journal. 2015;628195.
Crossref - Khehra M, Saini H, Sharma D, Chadha B, Chimni S. Decolorization of various azo dyes by bacterial consortium. Dyes Pigm. 2005;67(1):55-61.
Crossref - Bose P, Anitha R. Decolourization of Textile Dyes using Bacterial Consortium. Int J Sci Res Environ Sci. 2016;4(1):17-22.
Crossref - Neifar M, Sghaier I, Guembri M, et al. Recent advances in textile wastewater treatment using microbial consortia. J Text Engfash Technol. 2019;5(3):134-146.
Crossref - Lalnunhlimi S, Krishnaswamy V. Decolorization of azo dyes (Direct Blue 151 and Direct Red 31) by moderately alkaliphilic bacterial consortium. Braz J Microbiol. 2016;47(1):39-46.
Crossref - Meerbergen K, Willems KA, Dewil R, Van Impe J, Appels L, Lievens B. Isolation and screening of bacterial isolates from wastewater treatment plants to decolorize azo dyes. J Biosci Bioeng. 2018;125(4):448-456.
Crossref - Das A, Mishra S. Removal of textile dye reactive green-19 using bacterial consortium: Process optimization using response surface methodology and kinetics study. J Environ Chem Eng. 2017;5(1):612-627.
Crossref - Franciscon E, MendonCa D, Seber S, et al. Potential of a bacterial consortium to degrade azo dye Disperse Red 1 in a pilot scale anaerobic-aerobic reactor. Process Biochem. 2015;50(5):816-825.
Crossref - Eskandari F, Shahnavaz B, Mashreghi M. Optimization of complete RB-5 azo dye decolorization using novel cold-adapted and mesophilic bacterial consortia. J Environ Manage. 2019;241:91-98.
Crossref - Abubacker MN, Ayesha A. Decolourization of azo-dyes by bacterial consortium. Biosci Biotechnol Res Asia. 2011;8(2):741-746.
Crossref - John J, Dineshram R, Hemalatha KR, Dhassiah MP, Gopal D, Kumar A. Bio-Decolorization of Synthetic Dyes by a Halophilic Bacterium Salinivibrio sp. Front Microbiol. 2020;11:594011.
Crossref - Afrin S, Shuvo HR, Sultana B, et al. The degradation of textile industry dyes using the effective bacterial consortium. Heliyon. 2021;7(10):e08102.
Crossref - Jusoh N, Ruseli SNNM, Badri MF, Husin N, Hitam SMS. Biodecolourisation of Methyl Red Dye by bacterial-fungal consortium. Chemical Engineering Transactions. 2017;56:1537-1542.
Crossref - Das A. A Study on Evaluation of Indigenous Microbial Consortium for Enhanced Decolorization of Textile Azo Dyes and Feasibility for Simultaneous Bioelectricity Generation in A Microbial Fuel Cell. 2016. http://ethesis.nitrkl.ac.in/8427/1/2016-PhD-ADas-511CH110.pdf
- Cui D, Li G, Zhao D, Gu X, Wang C, Zhao M. Microbial community structures in mixed bacterial consortia for azo dye treatment under aerobic and anaerobic conditions. J Hazard Mater. 2012;221-222:185-192.
Crossref - Madhushika HG, Ariyadasa TU, Gunawardena SHP. Biological decolourization of textile industry wastewater by a developed bacterial consortium. Water Sci Technol. 2019;80(10):1910-1918.
Crossref - Nachiyar CV, Sunkar S, Kumar GN. Biodegradation of acid blue 113 containing textile effluent by constructed aerobic bacterial consortia: Optimization and mechanism. J Bioremediat Biodegrad. 2012;03(09).
Crossref - Sharma P, Singh L, Mehta J. COD reduction and colour removal of simulated textile mill wastewater by mixed bacterial consortium. Rasayan J Chem. 2010;3(4):731-735. http://www.rasayanjournal.co.in/vol-3/issue-4/21.pdf
- Tony BD, Goyal D, Khanna S. Decolorization of Direct Red 28 by mixed bacterial culture in an up-flow immobilized bioreactor. J Ind Microbiol Biotechnol. 2009;36(7):955-960.
Crossref - Parmar ND, Shukla SR. Decolourization of dye wastewater by microbial methods-A review. Indian Journal of Chemical Technology (IJCT). 2019;25(4):315-323. http://nopr.niscair.res.in/bitstream/123456789/44961/1/IJCT%2025%284%29%20315-323.pdf
- Ajaz M, Shakeel S, Rehman A. Microbial use for azo dye degradation-a strategy for dye bioremediation. Int Microbiol. 2020;23(2):149-159.
Crossref - Dong H, Guo T, Zhang W, et al. Biochemical characterization of a novel azoreductase from Streptomyces sp.: Application in eco-friendly decolorization of azo dye wastewater. Int J Biol Macromol. 2019;140:1037-1046.
Crossref - Sudha M, Saranya A, Selvakumar G, Sivakumar N. Microbial degradation of azo dyes: a review. Int J Curr Microbiol Appl Sci. 2014;3(2):670-690. https://www.researchgate.net/profile/Selvakumar-Gopal-3/publication/284548975_Microbial_degradation_of_Azo_Dyes_A_review/
links/5b46ef570f7e9b4637cde316/Microbial-degradation-of-Azo-Dyes-A-review.pdf - Rane A, Joshi SJ. Biodecolorization and biodegradation of dyes: A review. Open Biotechnol J. 2021;15(1):97-108.
Crossref - Kumar A, Kumar A, Singh R, et al. Genetically engineered bacteria for the degradation of dye and other organic compounds. Abatement of Environmental Pollutants. Elsevier. 2020:331-350.
Crossref - Jin RF, Zhou JT, Zhang AL, Wang J. Bioaugmentation of the decolorization rate of acid red GR by genetically engineered microorganism Escherichia coli JM109 (pGEX-AZR). World J Microbiol Biotechnol. 2008;24(1):23-29.
Crossref - Jin R, Yang H, Zhang A, Wang J, Liu G. Bioaugmentation on decolorization of C.I. Direct Blue 71 by using genetically engineered strain Escherichia coli JM109 (pGEX-AZR). J Hazard Mater. 2009;163(2-3):1123-1128.
Crossref - Liu GF, Zhou JT, Qu YY, Ma X. Decolorization of sulfonated azo dyes with two photosynthetic bacterial strains and a genetically engineered Escherichia coli strain. World J Microbiol Biotechnol. 2007;23(7):931-937.
Crossref - Eslami M, Amoozegar MA, Asad S. Isolation, cloning and characterization of an azoreductase from the halophilic bacterium Halomonas elongata. Int J Biol Macromol. 2016;85:111-116.
Crossref - Abbas A, Mushtaq A, Cheema AI, et al. Heterologous expression of azoreductase-encoding gene azrS of Bacillus sp. MR-1/2 for enhanced azo dye decolorization and wastewater treatment. Arch Microbiol. 2020;202(8):2135-2145.
Crossref - Mohandass S, Ragavan M, Gnanasekaran D, Lakshmanan U, Dharmar P, Saha SK. Overexpression of Cu/Zn superoxide dismutase (Cu/Zn SOD) in Synechococcus elongatus PCC 7942 for enhanced azo dye removal through hydrogen peroxide accumulation. Biology. 2021;10(12):1313.
Crossref - Sandhya S, Sarayu K, Uma B, Swaminathan K. Decolorizing kinetics of a recombinant Escherichia coli SS125 strain harboring azoreductase gene from Bacillus latrosporus RRK1. Bioresour Technol. 2008;99(7):2187-2191.
Crossref - Liang Y, Hou J, Liu Y, et al. Textile Dye Decolorizing Synechococcus PCC7942 Engineered With CotA Laccase. Front Bioeng Biotechnol. 2018;6.
Crossref - Chang JS, Lin CY. Decolorization kinetics of a recombinant Escherichia coli strain harboring azo-dye-decolorizing determinants from Rhodococcus sp. Biotechnol Lett. 2001;23(8):631-636.
Crossref - Tian F, Guo G, Zhang C, et al. Isolation, cloning and characterization of an azoreductase and the effect of salinity on its expression in a halophilic bacterium. Int J Biol Macromol. 2019;123:1062-1069.
Crossref - Zhang Y, Dong W, Lv Z, et al. Surface Display of Bacterial Laccase CotA on Escherichia coli Cells and its Application in Industrial Dye Decolorization. Mol Biotechnol. 2018;60(9):681-689.
Crossref - Jorgewad RM. Increasing efficiency of the dye degrading bacteria by Plasmid transfer method. SSR Institute of International Journal of Life Sciences. 2019;5(2):2218-2223.
Crossref - Rathod J, Dhebar S, Archana G. Efficient approach to enhance whole cell azo dye decolorization by heterologous overexpression of Enterococcus sp. L2 azoreductase (azoA) and Mycobacterium vaccaeformate dehydrogenase (fdh) in different bacterial systems. International Biodeterioration & Biodegradation. 2017;124:91-100.
Crossref
© The Author(s) 2023. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.