ISSN: 0973-7510
E-ISSN: 2581-690X
The profound impacts of global changes on biodiversity necessitate a more comprehensive documentation, particularly at the microscale level. To achieve precise and rapid insights into this unique diversity, the choice of an ideal species candidate is crucial. Neurospora crassa, a well-established organism in the field of biology, emerges as a promising candidate for this purpose. In our study, we explore the potential of the Carboxypeptidase A1 (CPA1) enzyme as a valuable tool for profiling global diversity. Our investigation has revealed that CPA1 possesses distinctive characteristics, notably its conserved solvent accessibility. This unique feature makes CPA1 an invaluable asset for microscale studies of global changes. The insights presented in our study serve as a practical blueprint, showcasing the application of structural biology in understanding diversity and global changes within microscale environments.
Neurospora crassa, Protease (Carboxypeptidase A1), Protein Modeling, Computational Analysis, Biodiversity
Genetic diversity regulated with comprehensive complex mechanisms in different living organisms.1,2 The diversity of genes and proteins deduced and regulated from environmental conditions.3,4 These regulations constructed the different structural biomacromolecules to fulfill living organisms’ essentials.5 Thus, evolution of specific traits should be traceable in the genetic codes of biomacromolecules as well as their structures.6,7 The cell and cell growth should adapt with different environmental stresses.8 Proteases as the important enzymes in the cells, can disclose this diversity.9,10
Most investigation on proteases was based on their needs in the market.11,12 Particularly microbial proteases that are used in many industries such as food processing and detergent.13 Many proteases investigated by researchers14, although protease (carboxypeptidase A1, CPA1) in Neurospora crassa gained inevitable and unavoidable position as it is from one easy culturing model fungus (N. crassa) besides its variety of approved functions in the cell.15-17 CPA1 involves in many cells function from protein maturation to immune response and reproduction.12,18
Diversity and climate change in global scale documented in many research articles.19,20 The changes are in many aspects of the living systems such as species interactions21, marines’ communities22, and populations22,23 as well as pest and disease shift24 besides evolutionary genetic25 and plasticity.26,27 These prospectives gained more attention in global scale.28,29 One important question of the many studies should be about the fingerprint changes of rising temperature and the ability of the earth to maintain the current biodiversity of plants and animals30 especially related to the shifts in the food supplies. However, the results and conclusion are mostly about the organism from different ecologically spread around the planet, thus the specific research on the organisms from smaller scale or even microscale31 can provide better picture. The research goals should be the effect of solar radiation and temperature on DNA repair32,33, thermotolerance34, drought resistance and different stresses on living organisms35,36 and their macromolecules. All the effects would have the direct and indirect changes on biomacromolecules specially proteins and enzymes expression and structures. Therefore, finding the microscale environment as well as ideal species and macromolecule/s to study these effects can really help our understanding on climate change and biodiversity.
To address this issue the N. crassa as the model microscopic fungus belong to the Sordariomycetes class was found around the tropical and subtropical regions17,37 and isolated from many environments. Thus, it can be good example to study the evolution based on the climate changes. Short life cycle and easiness in culture made it suitable organism for genetic study. Investigation of different biomacromolecules of N. crassa in genes and proteins structures can reveal different environmental stresses such as temperature and light more easily than another organism.38 Therefore, structural study of N. crassa macromolecules that can represent different environmental diversity can be very revealing, enlightening, and educational. Thus, the objective of this research paper is to scrutinize the protease (Carboxypeptidase A1) structure in N. crassa with the help of computational approaches. This information can help in designing the lab experiment with more meaningful approach. Meanwhile, it would be a good help to find the better link with environmental conditions and diversity of biomacromolecules.
The protease CPA1 was retrieved and annotated from the full genome of N. crassa in National Center for Biotechnology Information (NCBI). The specific physicochemical features and sequence analysis were done with the help of Swiss institute of bioinformatics-server (https://www.expasy.org/).39
The structural model was determined with homology modeling. The final models were evaluated with Ramachandran Map.40 Structural characterizations such as Secondary structure prediction and solvent accessible surface area were analyzed with Chou & Fasman secondary prediction and Fraczkiewicz and Braun’s method, respectively.41-45
Sequence alignment and phylogenetic tree were presented with Clustal Omega Multiple Sequence Alignment program [http://www.ebi.ac.uk/Tools/msa/clustalo/] and MultAlin server [http://multalin.toulouse.inra.fr/multalin/] and MEGA software version 4.0. Domain identification, subcellular localization and gene synteny were performed with the information from NCBI genomic database, respectively.46,47
Molecular weight of the CPA1 is around 45 KDa with neutral pH of isoelectric point (pI) (Table 1). The total of negative and positive charges residues in the structure of the CPA1 is equal. Aliphatic index (AI) and GRAVY showed this molecule is thermostable and slightly hydrophilic. The instability index indicated the stability of CPA1. The average estimated half time before degrading in the cell was around 20 to 30 hours. The extinction coefficient at 280 nm in water estimated to be 78520. The molecule consisted of high Ala, Gly, Ser and Thr residues (Table 2). The least residue is Cys that form two disulfide bonds (Cys250-Cys 274, Cys 326-Cys 361). There is signal peptide on the n-terminal sequence of CPA1.
Table (1):
Physiochemical features of N. crassa. Number of amino acids (NAA), molecular weight (MW), isoelectric point (pI), total number of negatively charged residues (Asp + Glu), total number of positively charged residues (Arg + Lys), aliphatic index (AI), grand average of hydropathicity (GRAVY)
Entry |
No. AA |
MW |
pI |
negatively AA (Asp + Glu) |
positive AA (Arg + Lys) |
Carbon |
Hydrogen |
Nitrogen |
Oxygen |
Sulfur |
Formula |
No. Atoms |
AI |
GRAVY |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
Q7S312 |
423 |
45899.58 |
7.11 |
38 |
38 |
2066 |
3148 |
550 |
620 |
9 |
C2066H3148N550O620S9 |
6393 |
76.45 |
-0.182 |
Table (2):
Amino acid compositions of the N. crassa CPA1
Ala (A) |
Arg (R) |
Asn (N) |
Asp (D) |
Cys (C) |
Gln (Q) |
Glu (E) |
Gly (G) |
His (H) |
Ile (I) |
Leu (L) |
Lys (K) |
Met (M) |
Phe (F) |
Pro (P) |
Ser (S) |
Thr (T) |
Trp (W) |
Tyr (Y) |
Val (V) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
54 |
15 |
21 |
20 |
4 |
12 |
18 |
36 |
9 |
19 |
30 |
23 |
5 |
19 |
16 |
32 |
32 |
8 |
23 |
27 |
12.80% |
3.50% |
5.00% |
4.70% |
0.90% |
2.80% |
4.30% |
8.50% |
2.10% |
4.50% |
7.10% |
5.40% |
1.20% |
4.50% |
3.80% |
7.60% |
7.60% |
1.90% |
5.40% |
6.40% |
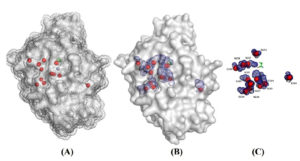
The 3d model structure provided with homology modeling with the procarboxypeptidase A (1PCa.1.A) template with more than 36 percent sequence identity and 92% coverage (from res24 to res 422) showed high percentage of helices followed by sheets secondary structure (Figure 1). The CPA1 is monomer with zinc ion. The evaluation of predicated model had QMEAN of -3.06 and Cβ=-2.14 with solvation around the -1.85 (Table 3).
Table (3):
N. crassa CPA1 model properties were predicated with homology modeling approach. N. crassa CPA1 information for the 3-D structures of as well as the secondary structure of CPA1
Entry |
Oligo state |
Ligand |
GMQE |
QMEAN |
Cβ |
Solvation |
Torsion |
Seq Identity |
Seq similarity |
Coverage |
Range |
QSQE |
Template |
Number of residues in favored region |
Number of residues in outlier region |
Helix (%) |
Sheet (%) |
Turn (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
Q7S312 |
Monomer |
Zinc |
0.71 |
-3.06 |
-2.14 |
-1.85 |
-2.22 |
36.60% |
0.39 |
0.92 |
27-422 |
0.00 |
1pca.1.A |
93.15% |
1.27% |
66 |
42 |
10 |
Figure 1. 3D model of CPA1 in solvent (A), surface and nucleus solvent accessible area (SASA) (B) and conserved residues (C). The data presented in angstrom (Å2)
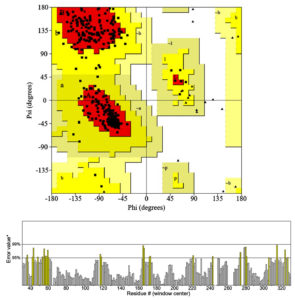
The model is highly accepted. Ramachandran map (Figure 2) indicated that more than 93 percent of residues were in highly acceptable position and only the 1.27% was considered as outsider (Table 3). The model was applied for the refinement with Galaxy Refine tool (http://galaxy.seoklab.org/cgi-bin/submit.cgi?type=REFINE), however the evaluation of the model didn’t change significantly, showed that homology modeling had significantly predicated the correct position of the CPA1 residues.
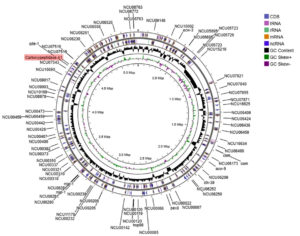
Protein-protein interaction showed that the CPA1 had interaction with serine peptidase, alpha-1,3-glucosyltransferase, endopeptidase K and glycoside hydrolase (1,4-galacrutonidase) (Figure 3). The whole genome of the N. crassa showed in the Figure 4 and the CPA1 was in the proximity with NCU07516 gene that is RING finger membrane protein.
Surface and nucleus accessibility showed residues VAL72, ILE73, LEU97, GLY178, ALA182, VAL190, ALA191, ASN224, GLY227, ASN241, GLY252, ASP254, ALA286, PRO321, GLY323, THR368, GLY369, ASP373, GLY396, ILE404, GLY408 had zero accessibility to the solvent (Table 4). These residues should be involved in the stability and conformational rigidity of the structure. On the other hand, the residues ARG27, LYS168, ARG341, ASP115, LYS298, TYR110, LYS 49, GLU86, LYS300 and ARG133 had the maximum accessibility to the solvent and considered as the functional residues.
Table (4):
Solvent accessibility of CPA1
| Surface atom | Buried atom | Apolar | Total SASA | Total SASA In/Out | Apolar In/Out | Backbone In/Out | Sidechain In/Out | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| nucleus | surface | nucleus | surface | nucleus | surface | nucleus | surface | ||||
| 1684 | 1359 | 9576.4 | 16122.86 | 1836.02 | 9508.03 | 1047.54 | 5725.47 | 573.63 | 1906.3 | 1262.41 | 7601.71 |
Finding the conserved residues in comparison with other CPAs showed that residues 181H, 183R, 184E, 224N, 226D, 227G, 240K, 257R, 258N, 280G, 373D and 410E were totally conserved beside that the residues 67G, 106G, 168K, 210S, 333N, 394A and 67G had high frequency compared to others. In the other positions all kind of the residues were seen and all of them exposed partially on the surface of the enzyme. It is interesting that conserved residues were all functional residues. Except for conserved residues, the tolerance of the residue exchange was acceptable in other positions of the structure. The maximum frequency was related to the positions 333N and 394A that can help to understand the most alternative residue in the structure. The residue exchange in other parts of the enzyme was observed however the frequency of them are not the same (Figure 1).
The scientific approach to find the interaction of biological diversity and climate condition is exploring based on the ideal model species and focusing on the diversity of major biological macromolecules in that species. Thus, structural biology of the biomacromolecule can be very suitable and informative specially with the help of computational analysis.48-52 Here N. crassa from the Sordariaceae family can be good example for the structural analysis of CPA1. This fungus was long time an ideal model for research in different aspects of molecular biology and genetics. Research on circadian rhythm-based physiological regulation53, RNA interference (RNAi) post-transcriptional gene silencing54,55, and DNA methylation-mediated epigenetic control56 have been done previously with this model organism. The good source of genetic sequences in public databases such as NCBI and many biological molecular tools and tractability as well as rapid culturing and single gene knock out collection made this fungus more attractable in filamentous ascomycete. The entire genome of N. crassa includes the seven chromosomes is available in the public data bases.57 This information can help to find the effect of the light and temperature58 particularly on CPA1 gene very easily during the 22 hours Lab work.59 Even complete growth cycle of N. crassa can easily observed and documented in less than one week. Additionally, the effect of the different environment on different life cycle time60 as well as gene annotation for specific genes would be easily documented.61
Structurally characterization of biomacromolecules provides great insight in defining the evolutionary and biodiversity changes during the time for specific species.62-64 Carboxypeptidases are the hydrolytic enzyme65 with the ability to cleave the c-terminal peptide bond of proteins and releasing free amino acids. They have many functional roles in cells such as degradation and modulation of intracellular proteins. They categorizing in three distinct groups including serine, metal and cysteine. Here in the structure of CPA1 the 3dmodel had zinc as the metal in the structure. CPA1 had two disulfide bonds in the structure that were also observed in animal cells polypeptides carboxypeptidases.66-71 Generally, peptidase or protease categorized on seven group based on the catalytic residues: serine, cysteine, threonine, aspartic, glutamic, asparagine and metalloproteases.12 The mechanism of action to cleave the peptide bond is by making the amino acid residue nucleophilic with catalytic triad. The histidine residue involved in the activation the serine, cysteine, or threonine as a nucleophile.
One important aspect for the investigation on CPA1 of the N. crassa is finding the changes in the active optimal pH. Furthermore, more information of specific zero accessibility to solvent residues identified in our research would help to understand the effect of different environmental stresses on evolution of this structure, additionally post translational modification of them would be informative in the specific environment. On the other hand, the residues with maximum accessibility to solvent could be very good target to find the environmental variability of these amino acids in microscale global changes. Combination of the solvent accessibility and the conserved residues could give the more information about the structural insight. This information (conserved and accessible solvent) in combination would help to find less and more tolerable part of the structure to examine for assessment of warming and global changes in macro and microscale. Meanwhile changing the amino acids would change many features of protein that provide great connection with environmental condition.
Reports on computational post translational Carboxypeptidase showed the slight modification of the molecules to performing its function.72-76 This post translational included the phosphorylation, O-glycosylation, acetylation in different sites of Serine, Threonine and Tyrosine.
Computational analysis of CPA1 from N. crassa, the model fungus presented. For future studies, the Lab isolating the new carboxypeptidase from this species in different climates (such as tropical or subtropical) and finding the SASA and comparing the results would provide better insight into the interaction of environmental stresses on this enzyme and generally biodiversity. Furthermore, finding the effect of light and temperature on CPA1 gene mutation and even in the post-translational modification of the structure would give better insight into global warming and biodiversity.
ACKNOWLEDGMENTS
The authors would like to thank the Faculty of Science and Technology, Suan Sunandha Rajabhat University and Mahidol University, Thailand, for their support.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
This work was supported by Suan Sunandha Rajabhat University (Grant no. 11773/2566).
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- Bervoets I, Charlier D. Diversity, versatility and complexity of bacterial gene regulation mechanisms: opportunities and drawbacks for applications in synthetic biology. FEMS Microbiol Rev. 2019;43(3):304-339.
Crossref - Strober B, Elorbany R, Rhodes K, et al. Dynamic genetic regulation of gene expression during cellular differentiation. Science. 2019;364(6447):1287-1290.
Crossref - Li Y, Kong D, Fu Y, Sussman MR, Wu H. The effect of developmental and environmental factors on secondary metabolites in medicinal plants. Plant Physiol Biochem. 2020;148:80-89.
Crossref - Kelly M. Adaptation to climate change through genetic accommodation and assimilation of plastic phenotypes. Philos Trans R Soc Lond B Biol Sci. 2019;374(1768):20180176.
Crossref - Breed MF, Harrison PA, Blyth C, et al. The potential of genomics for restoring ecosystems and biodiversity. Nat Rev Genet. 2019;20(10):615-628.
Crossref - Kulkarni P, Behal A, Mohanty A, Salgia R, Nedelcu AM, Uversky VN. Co-opting disorder into order: Intrinsically disordered proteins and the early evolution of complex multicellularity. Int J Biol Macromol. 2022;201:29-36.
Crossref - Prosdocimi F, de Farias ST. Origin of life: drawing the big picture. Prog Biophys Mol Biol. 2023;180-181:28-36.
Crossref - Gull A, Lone AA, Wani NUI. Biotic and abiotic stresses in plants. Abiotic and Biotic Stress in Plants. 2019:1-19.
Crossref - Dyer RP, Weiss GA. Making the cut with protease engineering. Cell Chem Biol. 2022;29(2):177-190.
Crossref - Razzaq A, Shamsi S, Ali A, et al. Microbial proteases applications. Front Bioeng Biotechnol. 2019;7:110.
Crossref - Naveed M, Nadeem F, Mehmood T, Bilal M, Anwar Z, Amjad F. Protease-a versatile and ecofriendly biocatalyst with multi-industrial applications: an updated review. Catal Lett. 2021;151:307-323.
Crossref - Gurumallesh P, Alagu K, Ramakrishnan B, Muthusamy S. A systematic reconsideration on proteases. Int J Biol Macromol. 2019;128:254-267.
Crossref - Abdal-Aziz SAA, Ali SM. Molecular characterization and differentiation of proteases isolated from different Aspergillus fungal species. Online J Biol Sci. 2021;21(1):69-119.
Crossref - Bond JS. Proteases: History, discovery, and roles in health and disease. J Biol Chem. 2019;294(5):1643-1651.
Crossref - Naureen U, Khosa AN, Mukhtar MA, Nabi F, Ahmed N, Saleem M. Genetic biodiversity and posttranslational modifications of protease serine endopeptidase in different strains of Sordaria fimicola. Biomed Res Int. 2023;2088988.
Crossref - Patel PK, Free SJ. The genetics and biochemistry of cell wall structure and synthesis in Neurospora crassa, a model filamentous fungus. Front Microbiol. 2019;10:2294.
Crossref - Gladieux P, De Bellis F, Hann-Soden C, Svedberg J, Johannesson H, Taylor JW. Neurospora from natural populations: Population genomics insights into the life history of a model microbial eukaryote. Methods Mol Biol.2020;2090:313-336.
Crossref - Ahmed T, Sun X, Udenigwe CC. Role of structural properties of bioactive peptides in their stability during simulated gastrointestinal digestion: a systematic review. Trends Food Sci Technol. 2022;120:265-273.
Crossref - Fordham DA, Jackson ST, Brown SC, et al. Using paleo-archives to safeguard biodiversity under climate change. Science. 2020;369(6507):eabc5654.
Crossref - Hutchins DA, Jansson JK, Remais JV, Rich VI, Singh BK, Trivedi P. Climate change microbiology-problems and perspectives. Nat Rev Microbiol. 2019;17(6):391-396.
Crossref - Boukal DS, Bideault A, Carreira BM, Sentis A. Species interactions under climate change: connecting kinetic effects of temperature on individuals to community dynamics. Curr Opin Insect Sci. 2019;35:88-95.
Crossref - Doney SC, Ruckelshaus M, Emmett Duffy J, et al. Climate change impacts on marine ecosystems. Ann Rev Mar Sci. 2012;4:11-37.
Crossref - Pauls SU, Nowak C, Bálint M, Pfenninger M. The impact of global climate change on genetic diversity within populations and species. Mol Ecol. 2013;22(4):925-946.
Crossref - Skendzic S, Zovko M, Zivkovic IP, Lesic V, Lemic D. The impact of climate change on agricultural insect pests. Insects. 2021;12(5):440.
Crossref - Waldvogel A-M, Feldmeyer B, Rolshausen G, et al. Evolutionary genomics can improve prediction of species’ responses to climate change. Evol Lett. 2020;4(1):4-18.
Crossref - Rodrigues YK, Beldade P. Thermal plasticity in insects’ response to climate change and to multifactorial environments. Front Ecol Evol. 2020;8:271.
Crossref - Sun B, Williams CM, Li T, et al. Higher metabolic plasticity in temperate compared to tropical lizards suggests increased resilience to climate change. Ecol Monogr. 2022;92(2):e1512.
Crossref - Sippel S, Meinshausen N, Fischer EM, Szekely E, Knutti R. Climate change now detectable from any single day of weather at global scale. Nat Clim Change. 2020;10(1):35-41.
Crossref - Gossling S, Humpe A. The global scale, distribution and growth of aviation: Implications for climate change. Global Environ Change. 2020;65:102194.
Crossref - Scheffers BR, Pecl G. Persecuting, protecting or ignoring biodiversity under climate change. Nat Clim Change. 2019;9(8):581-586.
Crossref - Banerjee A, Cornejo J, Bandopadhyay R. Emergent climate change impact throughout the world: call for “Microbiome Conservation” before it’s too late. Biodivers Conserv. 2020;29(1):345-348.
Crossref - Bornman JF, Barnes PW, Robson TM, et al. Linkages between stratospheric ozone, UV radiation and climate change and their implications for terrestrial ecosystems. Photochem Photobiol Sci. 2019;18(3):681-716.
Crossref - Neale R, Lucas R, Byrne S, et al. The effects of exposure to solar radiation on human health. Photochem Photobiol Sci. 2023;22:1011-1047.
Crossref - Gomez-Gras D, Linares C, de Caralt S, et al. Response diversity in Mediterranean coralligenous assemblages facing climate change: Insights from a multispecific thermotolerance experiment. Ecol Evol. 2019;9(7):4168-4180.
Crossref - Melton AE, Beck J, Galla SJ, et al. A draft genome provides hypotheses on drought tolerance in a keystone plant species in Western North America threatened by climate change. Ecol Evol. 2021;11(21):15417-15429.
Crossref - Ullah A, Nisar M, Ali H, et al. Drought tolerance improvement in plants: an endophytic bacterial approach. Appl Microbiol Biotechnol. 2019;103(18):7385-7397.
Crossref - Perkins DD, Turner BC. Neurospora from natural populations: toward the population biology of a haploid eukaryote. Exp Mycol. 1988;12(2):91-131.
Crossref - Aramayo R, Selker EU. Neurospora crassa, a model system for epigenetics research. Cold Spring Harb Perspect Biol. 2013;5(10):a017921.
Crossref - Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31(13):3784-8.
Crossref - Ramachandran G, Sasisekharan V. Conformation of polypeptides and proteins. Adv Protein Chem. 1968;23:283-437.
Crossref - Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16(22):10881-10890.
Crossref - Kumar TA. CFSSP: Chou and Fasman secondary structure prediction server. Wide spectr. 2013;1(9):15-19.
- Fraczkiewicz R, Braun W. Exact and efficient analytical calculation of the accessible surface areas and their gradients for macromolecules. J Comput Chem. 1998;19(3):319-333.
https://doi.org/10.1002/(SICI)1096-987X(199802)19:3<319::AID-JCC6>3.0.CO;2-W - Rost B, Sander C. Conservation and prediction of solvent accessibility in protein families. Proteins: Struct Funct Bioinform. 1994;20(3):216-226.
Crossref - Pettersen EF, Goddard TD, Huang CC, et al. UCSF Chimera-a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605-1612.
Crossref - Yu CS, Lin CJ, Hwang JK. Predicting subcellular localization of proteins for Gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Sci. 2004;13(5):1402-1406.
Crossref - Yu CS, Chen YC, Lu CH, Hwang JK. Prediction of protein subcellular localization. Proteins: Struct Funct Bioinform. 2006;64(3):643-651.
Crossref - Gruber J, Zawaira A, Saunders R, Barrett CP, Noble ME. Computational analyses of the surface properties of protein-protein interfaces. Acta Crystallogr Sect D Biol Crystallogr. 2007;63(1):50-57.
Crossref - Forster MJ. Molecular modelling in structural biology. Micron. 2002;33(4):365-384.
Crossref - Grasso D, Galderisi S, Santucci A, Bernini A. Pharmacological chaperones and protein conformational diseases: Approaches of computational structural biology. Int J Mol Sci. 2023;24(6):5819.
Crossref - Sraphet S, Javadi B. In silico analysis of Pseudomonas cellulose synthase A. Plant Cell Biotechnol Mol Biol. 2020;21(33-34):83-94.
- Javadi B. In silico characterization of lipase architectural structure in Rhizobium leguminosarum. Plant Cell Biotechnol Mol Biol. 2020;21(13-14):14-26.
- Loros JJ, Dunlap JC. Genetic and molecular analysis of circadian rhythms in neurospora. Annu Rev Physiol. 2001;63(1):757-794.
Crossref - Cogoni C, Macino G. Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature. 1999;399(6732):166-169.
Crossref - Cogoni C, Macino G. Posttranscriptional gene silencing in Neurospora by a RecQ DNA helicase. Sci. 1999;286(5448):2342-2344.
Crossref - Davis RH, Perkins DD. Neurospora: a model of model microbes. Nat Rev Genet. 2002;3(5):397-403.
Crossref - Kasuga T, Mannhaupt G, Glass NL. Relationship between phylogenetic distribution and genomic features in Neurospora crassa. PLoS One. 2009;4(4):e5286.
Crossref - Diernfellner AC, Schafmeier T, Merrow MW, Brunner M. Molecular mechanism of temperature sensing by the circadian clock of Neurospora crassa. Genes Dev. 2005;19(17):1968-1973.
Crossref - Gaba A, Jacobson A, Sachs MS. Ribosome occupancy of the yeast CPA1 upstream open reading frame termination codon modulates nonsense-mediated mRNA decay. Mol Cell. 2005;20(3):449-460.
Crossref - Graham JK, Smith ML, Simons AM. Experimental evolution of bet hedging under manipulated environmental uncertainty in Neurospora crassa. Proc R Soc Lond B Biol Sci. 2014;281(1787):20140706.
Crossref - Carrillo AJ, Schacht P, Cabrera IE, et al. Functional profiling of transcription factor genes in Neurospora crassa. G3 (Bethsda). 2017;7(9):2945-2956.
Crossref - Wang R, Arioka M. Functional analyses of xylanolytic enzymes involved in xylan degradation and utilization in Neurospora crassa. Int J Biol Macromol. 2021;169(1):302-310.
Crossref - Sraphet S, Javadi B. Application of hierarchical clustering to analyze solvent-accessible surface area patterns in Amycolatopsis lipases. Biology. 2022;11(5):652.
Crossref - Sraphet S, Javadi B. Computational characterizations of GDP-mannose 4, 6-dehydratase (NoeL) Rhizobial proteins. Curr Genet. 2021;67(5):769-784.
Crossref - Lai X, Tang J, ElSayed ME. Recent advances in proteolytic stability for peptide, protein, and antibody drug discovery. Expert Opin Drug Discov. 2021;16(12):1467-1482.
Crossref - Breddam K. Serine carboxypeptidases. a review. Carlsberg Res Commun. 1986;51:83-128.
Crossref - Song P, Xu W, Zhang Y, et al. A new carboxypeptidase from Aspergillus niger with good thermostability, pH stability and broad substrate specificity. Sci Rep. 2021;11(1):18745.
Crossref - Bonten EJ, Galjart NJ, Willemsen R, Usmany M, Vlak JM, d’Azzo A. Lysosomal Protective Protein/Cathepsin A: role of the “linker” domain in catalytic activation.
J Biol Chem. 1995;270(44):26441-26445.
Crossref - Jung G, Ueno H, Hayashi R. Carboxypeptidase Y: structural basis for protein sorting and catalytic triad. J Biochem. 1999;126(1):1-6.
Crossref - Endrizzi JA, Breddam K, Remington SJ. 2.8-ANG. structure of yeast serine carboxypeptidase. Biochem. 1994;33(37):11106-11120.
Crossref - Ejalonibu MA, Ogundare SA, Elrashedy AA, et al. Drug discovery for Mycobacterium tuberculosis using structure-based computer-aided drug design approach. Int J Mol Sci. 2021;22(24):13259.
Crossref - Saleem M, Lamb BC, Nevo E. Inherited differences in crossing over and gene conversion frequencies between wild strains of Sordaria fimicola from “Evolution Canyon”. Genetics. 2001;159(4):1573-1593.
Crossref - Ishfaq M, Mahmood N, Nasir IA, Saleem M. Molecular and biochemical screening of local Aspergillus niger strains efficient in catalase and laccase enzyme production. Int J Agric Biol. 2014;16(1).
- Arif R, Bukhari SH, Ishfaq M, Shahid MG, Lee SF, Saleem M. Genetic variation and post-translational modifications of cytochrome c oxidase-1 (COX1) in different strains of Sordaria fimicola. Int J Agric Biol. 2019;21:1055-1062.
Crossref - Bukhari SH, Mobeen I, Naureen U, et al. Analysis of genetic polymorphisms and post translational modifications of cytochrome C-1 in Sordaria fimicola. Int J Agric Biol. 2020;23(3):675-680.
Crossref - Mobeen I, Arif R, Rasheed A, Akram F, Shahid MG, Saleem M. Genetic and post-translational modification analysis of translational associated protein RKM4 in Sordaria fimicola. Int J Agric Biol. 2020;23(5):935-942.
Crossref
© The Author(s) 2024. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.