ISSN: 0973-7510
E-ISSN: 2581-690X
Heme, a Fe-centred hexacoordinated organometallic compound mainly present in blood proteins, acts as a major source of iron (Fe2+) for various hemolytic microorganisms. This makes blood an essential mediumsupplement for characterizing such organisms. Considering the increasing blood demand for this purpose, we are proposing an alternative approach for hemolytic characterization of bacteria using plant derivative. For the first time, we present the kinetic model of growth of Staphylococcus aureus, a beta-hemolytic, gram-positive rod-shaped bacterium, and subsequent heme uptake at an optimized concentration of crude proteins of Beta vulgaris (red beetroot). In this paper, we have determined the heme content of the beetroot juice and demonstrated the ability of S. aureus to uptake heme from beetroot protein solution. According to the determined growth kinetics, the doubling time of the bacterium in the beetroot protein supplemented medium was 25 minutes. The heme uptake analysis showed a zeroth order kinetics with a constant decline rate of 0.19 µM h-1.
Plant heme, 3R’s, Beetroot, Hemolytic characterization, Heme uptake kinetics, Growth kinetics.
Iron-centred hexacoordinated heme complex serves as a major co-factor in numerous enzymes that regulate the metabolic pathways and helps to sustain life. Microorganisms that lack heme biosynthetic pathways depend on exogenous sources to fulfill their heme or iron requirement. Some micro-organisms are capable of scavenging iron from heme proteins, such as hemoglobin. These organisms disintegrate the membrane of the RBCs to release the hemoglobins. These non-bounded hemoglobins are easily accessible to the microbes to chelate iron via different mechanisms (Mayer, 1996).
To demonstrate the hemolytic activity of the microorganisms, blood agar plating is a traditional method, which requires supplementation of 5-10% blood. However, blood is not the sole source of heme/iron for hemolytic bacteria. A diversity of microbes has the capacity to scavenge iron from various other heme proteins. Property of the beta hemolytic bacteria to utilize heme from various animal hemeproteins, such as transferrin, lactoferrin, heme, hemoglobin, etc. (Mietzner and Morse, 1994; Lee, 1995) redirects to the possibility that those bacteria can also uptake iron from various plant heme-proteins.
Furthermore, the increasing blood demand is fulfilled by harvesting 15-20% of the total blood volume from several animals (like goats, sheep, pigs, etc.; Gratten et al, 1994; Animal Welfare Advisory Committee NZ, 1996; Anand et al., 2000; Russell et al., 2006) on a large scale, annually. The annual increase in this data is a matter of high ethical concern. The use of blood incurs various drawbacks in culture, such as expensiveness; contamination of viruses, Mycoplasma, etc.; batch variation (DeGraw et al, 1979; Hill, 1984; Whitney et al., 2003); difficulty in sterilization; limited shelf-life; high demand to supply ratio; safety-, regulatory- and ethical- issues (Malikides et al., 2000; Malikides et al, 2001); cause damage to the environment; incompatibility of blood with bacterial growth (Krumwiede and Kuttner, 1938); source of development of multi-drug resistant (MDR) strains; etc. (Suman et al., 2015). It seems paradoxical to use blood during in vitro studies in favor of 3R’s principle. Considering these shortcomings of supplementing blood in basal medium for bacteria, it becomes necessary to promote the 3R’s principle in a new way (Russell and Burch, 1959; Balls et al., 1995).In this paper, we are proposing an alternate approach to demonstrate the ability of hemolytic bacteria to uptake heme from plant derivatives.
Here, we are demonstrating the same using proteins from Beta vulgaris (red beetroot; family Amaranthaceae), which is usually grown for the commercial and feeding purpose. It has been demonstrated that non-symbiotic hemoglobin is predominantly expressed in different parts of beetroot, including leaves and roots and has structural similarity to neuroglobin, a hexacoordinated Fe-centred hemoglobin present in thebrain (Leiva-Eriksson et al., 2014).
In progression toward alternatives to the use of animals or animal products in experimentation, this work opens a wide scope and novel trend for researchers to use and further evolve the technique of characterizing hemolytic bacteria using plant derivatives.
Plant material
Crude juice and proteins isolated from the crude juice of red beetroot (Beta vulgaris L., family – Amaranthaceae) was used in the study.
Collection of beetroot
From the botanical garden in Jalandhar, Punjab, 500 grams of fresh and dark red beetroot without any observable sign of infection, was bought and kept in water for overnight at 4°C.
Preparation of beetroot juice
After washing twice with running tap water and distilled water,200g red beetroot was grated mechanically and ground using a domestic electric grinder at low to medium speed for 2-3 minutes repeatedly for 5-6 times, meanwhile refrigerating it at 4°C for 5 minutes post-operation. The fluidized slurry, thus obtained, was strained through a muslin cloth to remove the large solid materials. The filtrate was, then, kept at 4°C for overnight, to precipitate cold-insoluble materials.The filtrate was then centrifuged at 9400×g for 20 minutes at 4°C to sediment the suspending particles. The supernatant was filtered using Whatman filter paper (porosity 1.5 µm). The filtrate was sterilized using vacuum-filter (porosity 0.45 µm) and 0.22 µm syringe filter, under aseptic condition, collected and stored at 4°C.
Media preparation
Mannitol salt agar was prepared by dissolving 110g HiVegTM Mannitol salt agar (HiMedia) in 1000ml distilled water.
Nutrient broth (w/ 7.5% NaCl) was prepared by dissolving the ingredients listed in table 1:
Table (1):
Composition of Nutrient broth (w/ 7.5% NaCl).
Ingredient |
Amount (g/L) |
|---|---|
Bacteriological peptone |
5.00 |
Sodium Chloride |
75.00 |
Beef extract |
1.50 |
Yeast extract |
1.50 |
Final pH (at 25°C) 7.4 ± 0.2
Bacterium used
The strain of Staphylococcus aureus (MTCC 740), was used for this study. It was maintained on freshly prepared Mannitol Salt Agar plate in triplicate and incubated at 37°C for 48 hours. Then, the cultures were stored at 4°C, till further use.
Clonal growth of S. aureuson beetroot juice agar
Beetroot juice agar consisted autoclaved 1% (w/v) agar solution (Agar Powder; HiMedia) and beetroot juice (10%, v/v) which was supplemented just before solidification of the agar solution. The plates were inoculated with the maintained S. aureus culture and incubated for 24h at 37°C.
Quantification of heme in beetroot juice
The concentration of heme in beetroot juice was determined using QuantiChrom™ Heme Assay Kit (DIHM-250; Bioassay systems) in a 96-well microtiter plate, following the procedure provided along with the kit. (Supplementary file 1, section A).
Purification of beetroot juice proteins
The protein content in the beetroot was purified using ammonium sulfate precipitation method (Wingfield, 2001). Solid (NH4)2SO4(66.82g) was added in 100ml of beetroot juice to achieve 95% saturation at 4°C(http://encorbio.com/protocols/AM-SO4.htm)and kept on amagnetic stirrer at slowstirring rate for 6h. pH of the solution was maintained at 7.0 using 10N NaOH. The protein suspension, thus obtained, was centrifuged at 9400×g for 15 min. and the pellet was dissolved in 20ml of 0.1M PBS (pH 7.0). The solution was dialyzed against 0.1M PBS (pH 7.0) using a dialysis membrane (LA 387-5MT; HiMedia) at 4°C.Post-dialysis the solution was collected in a sterile falcon tube and stored at 4°C.
Protein estimation assay
The concentration of the proteins in the solution from the beetroot juice was estimated by Lowry’s method. (Supplementary file 1, section B).
Growth of S. aureus at different concentration of beetroot protein
Two different sets of nutrient broth (table 1) were taken. The first set (experimental medium) was supplemented with 7 different concentrations (i.e. 60, 30, 15, 7.5, 3.75, 1.88 and 0.94 µg/ml) of beetroot proteins. The other set (control medium) was kept as such. As shown in figure 1, in separate 7 rows of the first 6 consecutive columns of a 96-well microtiter plate (12 columns x 8 rows), 100µl of the experimental medium was taken. In the first 6 columns of the last row, 100µl of the control medium was taken. In all the rows of first 3 columns,100µl of the bacterial culture (OD=0.3) was inoculated. This diluted the protein concentration in the experimental medium by half (i.e. 30, 15, 7.5, 3.75, 1.88, 0.94, 0.47 µg/ml, respectively). In all the rows of the next three columns, 100µl nutrient broth was added to equalize the total volume (200µl) and the protein concentrations in the experimental and the control medium. The plate was sealed and incubated at 37°C for 24h followed by spectrophotometry (at 600nm wavelength).
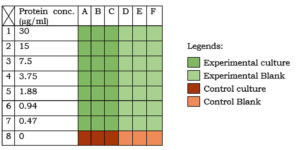
Fig. 1:Experimentation in microtiter plate. Different colours indicate different sets (as described in the legend). Protein conc. in the medium is indicated on the left-hand side of the table.
The percentage relative growth was determined using the equation: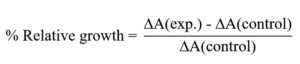 Where,
Where,
ΔA(exp.) = OD of experimental culture – OD of experimental blank
ΔA(control) = OD of control culture – OD of control blank
Here, the term percentage relative growth indicates the difference between the bacterial concentrations (as optical density at 600nm) in the experimental and the control sets with respect to the control set. The protein concentration at which maximum relative growth was observed was considered as the optimum concentration and used in further experimentation.
Evaluation of growth kinetics of S. aureus
In two flasks, each containing 50ml nutrient broth (table 1) enriched with 6µg/ml beetroot protein (optimized conc.), 1ml of prepared inoculum of S. aureus culture (OD = 0.2±0.01) was inoculated. From each flask, 5ml culture was collected and OD (at 600nm wavelength) was determined against the same un-inoculated enriched medium composition. The observed OD was 0.004. At a regular interval of 1 hour, for next 8 hours, 1ml sample from each flask was collected in separate sterile eppendorf tubes (2ml) and centrifuged at 2500rpm for 10 min. The supernatants were collected in separate sterile eppendorf tubes and kept at 4oC to determine the remaining heme concentration. The pellets (cells) were washed twice with 7.5% NaCl solution, re-suspended in 1ml solution of 7.5% NaCl and OD at 600nm wavelength was determined against sterile 7.5% NaCl solution.
The cell concentration was calculated from OD using the following equation (Rasigade et al., 2011) and the growth curve of OD or cell conc. (Y-axis) vs. time (X-axis) was plotted:
Cell conc. (cells/ml) = OD (600nm)
× 109 × Dilution factor
Evaluation of heme utilization kinetics for S. aureus
The concentration of heme in the stored culture supernatant was determined as mentioned in section 2.7 (Supplementary file 1, A). The curve between the heme conc. (µM; Y-axis) and time (h; X-axis) was plotted. The curve showed the heme conc. that remained in the culture with respect to time.
Statistical analysis
All the statistical analysis (mean, standard deviation, t-test, correlation, etc.) were done using GraphPad Prism v5.0 for Windows.
Yield of beetroot juice
Processing of beetroot yielded 0.5 liters of juice per kilogram of beetroot.
Clonal growth of S. aureus on beetroot juice agar
On beetroot juice agar, good growth (>70 cfu) was observed (Fig. 2). Promotion of clonal growth of S. aureus in beetroot juice agar revealed that the juice contains sufficient nutrients required by the bacterium. This strengthened our hypothesis that the proteins of beetroot juice may also do the same.
Quantification of heme in beetroot juice
The estimated molar concentration of heme (Mol. Wt. = 616.487 g/mol) in the beetroot juice was 177.11µM, equivalent to 109.186µg/ml (for estimation and calculation refer section C of Supplementary file 1).
Protein estimation
Based on the plotted BSA standard curve (y = 0.0024x + 0.0006; R2 = 0.9997; y = absorbance at 750nm, x = conc. of BSA (µg/ml)), the concentration of protein in the solution was calculated. The estimated concentration of protein in the solution was 600µg/ml. Therefore, the amount of the protein in the collected 35ml solution was 21mg.
Effect of heme proteins on growth of S. aureus
As shown in figure 3, the maximum relative growth (57%) of the bacteria was observed at 7.5µg/ml protein conc., which indicated that the growth in the nutrient broth enriched with 7.5µg/ml beetroot protein was greater than that in the nutrient broth by 57%. Hence, we considered 6µg/ml as the optimized concentration.

Fig. 3: Percentage relative growth of S. aureus at different conc. of beetroot proteins. Maximum growth of 57% was observed at 7.5µg/ml.
Growth kinetics of S. aureus
The growth curve of S. aureus in the nutrient broth enriched with 6µg/ml beetroot protein is shown in figure 4. The statistical analysis of the growth kinetics showed a specific growth rate (µ) of 1.767±0.017 at 95% confidence interval, indicating a doubling time of 24 minutes.

Fig. 4: Growth curve of S. aureus in nutrient medium enriched with 6µg/ml beetroot protein on a semi-log scale. Y-axis represents the logarithmic values of the cell conc. (cells/ml).
Heme utilization kinetics
The heme utilization curve of S. aureus(figure 5) shows the decreasing molar concentration of heme in the medium with respect to time. The decrease in the hemeconc. in the medium is equal to the heme utilized by the cells. Thus, the rate of decrease in the hemeconc. is equal to the rate of heme utilized by the bacteria. Statistical analysis of the curve revealed that heme utilization by the bacteria is azeroth order kinetics with a constant rate of 0.19 ± 0.0042 µMh–

Fig. 5:Heme uptake kinetics for S. aureus. The curve represents decreasing conc. of heme in the culture with respect to time.
The logarithmic increase in the bacterial concentration and the corresponding linear decrease in the heme concentration indicate that the heme requirement of S. aureus decreases exponentially with increase in the cell concentration.
In various oxidoreductase proteins (Class I enzymes), heme is a prosthetic group whose central metal has high affinity for oxygen. Being a compound, its structure remains same in almost all the heme-proteins. In plants, heme is present in several proteins, like chlorophyll, cytochrome c oxidase, peroxidase, catalase and plant haemoglobins (Dekock et al., 1960). The presence of non-symbiotic hemoglobin in the beetroot and its similarity with the human neuroglobin (Leiva-Eriksson et al., 2014) directed us to determine the heme concentration in the beetroot juice.
After a successful attempt of culturing the bacteria on the beetroot agar medium, when tested with a wide range of beetroot protein concentration in the nutrient broth medium, the cells showed elevated level of growth in comparison to that in nutrient broth itself. Maximum growth was observed at concentration 7.5 µg/ml. This was considered as optimum concentration for growth of S. aureus.
Determination of growth kinetics at optimized concentration of beetroot protein revealed the doubling time (25 minutes) of S. aureus in the enriched medium. Further, determination of heme uptake revealed that decline in the heme concentration followed zeroth order kinetics and was constant throughout the process for at least 8 hours. This explained that with increase in the population density, the rate of heme uptake dropped. This can be concluded as the cells require more heme at early stage (lag-phage and early log-phase) of their growth cycle. The studies on the biochemical pathways and molecular mechanisms of S. aureus are to be carried out for uptake of heme from plant source
This study confirms our hypothesis (Suman et al., 2015) that apart from blood, the hemolytic micro-organism, S. aureus, can also uptake heme from plant derivatives. However, this study needs to be done on other hemolytic micro-organisms. This study also opens a window to develop a blood-free, plant-based system for characterizing the haemolytic activity of the micro-organisms.
- Anand, C.R., Gordon, H., Shaw, H., Fonseca, K., Olsen, M.. Pig and goat blood as substitutes for sheep blood in blood-supplemented agar media. J. Clin. Microbiol., 2000; 38: 591-594.
- Animal Welfare Advisory Committee NZ.. Guidelines for the welfare of livestock from which blood is commercially harvested for commercial and research purposes, Biosecurity New Zealand, NZ, 1996.
- Balls, M., Goldberg, A. M., Fentem, J. H., Broadhead, C. L., Burch, R. L., et al.. The three Rs: the way forward. The report and recommendations of ECVAM Workshop 11. ATLA., 1995; 23: 838-866.
- deGraw, W. A., Kern, M. D., King, J. R.. Seasonal changes in the blood composition of captive and free-living White-crowned sparrows. J. Comp. Physiol., 1979; 129: 151-162.
- Gratten, M., Battistutta, D., Torzillo, P., Dixon,
J. Manning, K. Comparison of goat and horse blood as culture medium supplements for isolation and identification of Haemophilus influenzae and Streptococcus pneumoniae for upper respiratory tract secretions. J. Clin. Microbiol., 1994.; 32: 2871–2872. - Hille, S.. The effect of environmental and endogenous factors on blood constituents of rainbow trout (Salmo gairdneri)—I. Food content of stomach and intestine. Comp. Biochem. Phys. A., 1984; 77: 311-314.
- Krumwiede, E., Kuttner, A. G.. A growth inhibitory substance for the influenza group of organisms in the blood of various animal species. The use of the blood of various animals as a selective medium for the detection of haemolytic Streptococci in throat cultures. J. Exp. Med., 1938; 67: 429-441.
- Lee, C. B.. Quelling the red menace: heme capture by bacteria. Mol. Microbiol., 1995; 18: 383–390.
- Leiva-Eriksson, N., Pin, P. A., Kraft, T., Dohm, J. C., Minoche, A. E., Himmelbauer, H., Bulow, L.. Differential expression patterns of non-symbiotic hemoglobins in sugar beet (Beta vulgaris ssp. vulgaris). Plant Cell Physiol., 2014; 55(4): 834-844.
- Malikides, N., Mollison, P. J., Reid, S. W. J., Murray M. Haematological responses of repeated large volume blood collection in the horse. Res. Vet. Sci., . 2000; 68: 275–278.
- Malikides, N., Hodgson, J. L., Rose, R. J., Hodgson D.R. Cardiovascular, haematological and biochemical responses after large volume blood collection in horses. Vet. J., 2001.;162: 44-55.
- Meyer, J. M., Neely, A., Stintzi, A., Georges, C., Holder, I. A. Pyoverdin is essential for virulence of Pseudomonas aeruginosa. Infect. Immun., 1996; 64: 518–523.
- Mietzner, T. A., Morse, S. A.. The role of iron-binding proteins in the survival of pathogenic bacteria. Annu. Rev. Nutr., 1994; 14: 471–493.
- Rasigade, J. P., Moulay, A., Lhoste, Y., Tristan, A., Bes, M., Vandenesch, F., Etienne, J., Lina, G., Laurent, F. Dumitrescu, O. Impact of sub-inhibitory antibiotics on fibronectin-mediated host cell adhesion and invasion by Staphylococcus aureus. BMC Microbiol., 2011; 11: 263. Doi:10.1186/1471-2180-11-263.
- Russell, F.M., Biribo, S. S. N., Selvaraj, G., Oppedisano, F., Warren, S., Seduadua, A., Mulholland, E. K., Carapetis, J. R. As a bacterial culture medium, citrated sheep blood agar is a practical alternative to citrated human blood agar in laboratories of developing countries. J. Clin. Microbiol., 2006.; 44: 3346-3351.
- Russell W. M. S., Burch R. L.. The principles of humane experimental technique. (ed. Russell W. M. S.) (Methuen, London,)., 1959.
- Suman A., Singh A. Plant heme: Future approach for characterizing haemolytic bacteria. Res. J. Pharm. Biol. Chem. Sci., 2015; 6: 826-830.
- Whitney, A. R. Diehn, M., Popper, S. J., Alizadeh, A. A., Boldrick, J. C., Relman, D. A., Brown, P. O.. Individuality and variation in gene expression patterns in human blood. Proc. Natl. Acad. Sci. USA., 2003; 100: 1896-1901.
- Wingfield, P. T.. Protein precipitation using ammonium sulfate. Curr. Protoc. Protein Sci. 2016; Appendix 3F.Doi:10.1002/0471140864.psa03fs84.
© The Author(s) 2017. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.