ISSN: 0973-7510
E-ISSN: 2581-690X
The mounting disquiets about the usage of precarious chemicals in the textile industry have steered to the development of eco-friendly and biological methods of fiber processing in the ever-escalating horizon of textile fibers. The current study targets the isolation, identification, and screening of lignin-degrading bacteria from termite gut microflora which could be employed in the textile trade, especially in coir industries for developing a biological method for softening coir fibers. Based on the morphology and taxonomic keys, termites used in the study were identified as Odontotermes obesus. The bacteria isolated from the termite gut having lignolytic activity were picked by using the methylene blue dye decolorizing method. The same was confirmed by using tannic acid. The isolates were then identified as Kosakonia oryzendophytica and Pseudomonas chengduensis by 16s rRNA sequencing. The isolates were further checked for their ability to produce extracellular lignolytic enzymes. The enzyme concentration was found to be significantly higher in the medium containing the microbial consortium than in those with the individual cultures. The consortium filtrate has MnP activity of 41.6 U/mL, LiP activity of 114.3 U/mL, and laccase activity of 61.85 U/mL at 72 hours of incubation. It was found that the enzyme activities were increased considerably until 72 hours of incubation but showed an insignificant increase at 96 hours.
Lignolytic, Peroxidases, Laccase, Facultative Anaerobe
Lignocellulose is a most profuse and low-cost renewable biological resource and signifies a chief source of renewable biotic material, on the other hand it is challenging to be degraded due to its multifaceted assembly.1 The sale and export of lignocellulosic fibres contribute significantly to the income and food security of poor farmers and those working in fibre processing and marketing in many developing nations.2 The physical parameters and characteristics of natural lignocellulosic fibers hinder the processing methods. The outsized and variable diameter, high microfibrillar angle, hemicellulose and greater lignin content have been found responsible for its poor power-driven reinforcement. Chemical treatments have many disadvantages as it releases harmful compounds to the environment, and may deteriorate the strength of natural fibers with time.3 Researches are underway to discover eco-friendly approaches to attain softened natural fibers by removing lignin which plays the foremost role in its tough nature. The ability of gut bacteria in termites to disrupt the plant cell wall polysaccharide is closely 30% – 40% superior to the large herbivores. Nonetheless, limited studies have been performed on the identification and screening of lignolytic bacteria from the gut of termites.
The most important lignolytic enzymes are laccases (E.C. 1.10.3.2) and peroxidases: manganese peroxidase (E.C. 1.11.1.13) and lignin peroxidase (E.C. 1.11.1.14).4 A diverse array of microbes particularly, fungi and bacteria, including actinomycetes, are capable to decompose lignin, however, fungi have the highest degrading ability.5
The termites are one of the greatest significant lignocellulose decomposers in the natural biota predominantly in the tropics. Termites, which are proficient lignocellulose degraders, flourish on plant debris and contribute to carbon mineralization, particularly in tropical and subtropical regions.6-9 The gut microbial community comprises all three domains of organisms: Bacteria, Archaea, and Eukaryotes (protists). Fairly little is found in the foregut and midgut, whereas abundant microbiota is found in the hindgut.10,11 There are several types of bacteria with diverse utilities in the termite’s gut that have been isolated and identified. They were hemicellulose degrading bacteria12 lignolytic bacteria,13 cellulolytic bacteria,14 aromatics degrading bacteria,15 and nitrogen-fixing bacteria.16 The most plentiful bacteria which have been recognized from both higher and lower termites belong to the species of strict or facultative anaerobes. The role of the facultative anaerobes may be to scavenge the oxygen that can infuse into the gut, effectually keeping the gut anaerobic.17 The present study aims at isolation, identification, and preliminary screening of lignin-degrading microbes from the gut of termites, Odontotermes obesus.
Collection and Identification of Termites
Soldier termites were collected from their nest from the backyard of the Central Coir Research Institute and were checked for their identity with the help of Trinocular Stereo Zoom Microscope (dissection microscope), LSZM1. The Identification of termites was done following the schemes proposed by Roonwal and Chhotani18 and Chhotani.19
Bacterial Isolation
Worker termites were collected from their nests from the backyard of the Central Coir Research Institute campus and surface sterilized using 70% ethanol followed by mashing with mortar and pestle by adding one mL normal saline and subjected to serial dilution. The ground mixture was transferred to 99 ml Berg minimal media containing the following ingredients per litre: 2 g NaNO3, 0.5 g K2HPO4, 0.02 g MgSO4:7H2O, 0.02 g MnSO4:7H2O, 0.02 g FeSO4:7H2O, and 0.5 g CaCl2:2H2O with 1% kraft lignin as the sole carbon source and subjected to serial dilution. Nutrient agar plates were prepared with the addition of 1% kraft lignin as sole source of carbon and 25 mg/L methylene blue as an indicator dye for picking out the lignolytic microbes exhibiting zone of decolourization around the bacterial colonies.20,21 Plates were inoculated with dilutions of 10-3, 10-4, 10-5, 10-6, and 10-7 respectively, and incubated at 37°C for 48 hours. The colonies with clearance zone were isolated and streaked over fresh plates and incubated at 37°C for 48 hours. The lignolytic activity was further confirmed by using tannic acid in the culture medium as a substitute for lignin.22
Identification of the Isolated Bacterial Strains
The isolated DNA sample was amplified by performing PCR using 16S Forward (AGAGTTTGATCCTGGCTCAG) and 16S Reverse (GGTTACCTTGTTACGACTT ) primers. 50 μl PCR mixture consisted of 1 μl template DNA, 5 μl Taq reaction buffer (10X), 1 μl 200 μM dNTPs, 0.25 μl of Taq DNA polymerase, 1 μl of 10 μM concentration of forward and reverse primers. The reaction mixture was finally made upto 50 μl with nuclease free water. The cycling conditions were; Initial denaturation at 95°C for 30 seconds, 30 cycles consisting of template denaturation at 95°C for 15-30 seconds, primer annealing at 45-68°C for 15-60 seconds, primer extension at 68°C for one minute per kb and a final extension at 72°C for 5 minutes. The amplicon was then electrophoresed in 2% SYBR SAFE agarose gel at 85 volts and visualized under UV and the concentration of the same was checked using Nanodrop ND 8000. The amplicon was further purified with the help of PureLink purification column (Invitrogen) and sequencing was done by sanger sequencing method by using forward and reverse primers in the ABI 3730xl cycle sequencer. Forward and reverse sequences were assembled and contig was generated after trimming the low-quality bases. The sequence analysis was performed using BLAST, the Bioinformatic tool of NCBI. Based on the maximum identity score first, a few sequences were nominated and aligned by multiple sequence alignment software ClustalO. The dendrogram was constructed. All sequences obtained were submitted to the GenBank nucleotide database (GenBank Accn No. OM943747, OM943748).
Enzymatic Assays
To perform the lignolytic enzyme assays, 5 mL of the culture media was drawn out at 24, 48, 72, and 96 hours respectively. The drawn cultures were then centrifuged at 12000 rpm to obtain the extracellular enzymes present in the medium. The supernatant (crude enzyme) obtained after centrifugation was taken for testing the presence of lignolytic enzymes produced by the microbes.
Lignin Peroxidase (E.C. 1.11.1.14) Assay
Lignin peroxidase enzyme activity was determined by observing the oxidation of azure B dye in the presence of H2O2 (100µM). The reaction mixture taken for the test comprised 125 mM of 1 mL sodium tartarates buffer (pH 3.0), 500 of 2 mM H2O2, 500 µL of 160 µM Azure B and 0.5 mL of the enzyme extract. The absorbance was read at 310 nm after the reaction is being initiated by the addition of 0.5 mL of H2O2. One unit of enzyme activity is equivalent to an decrease in absorbance of 0.1 units per minute per mL.23
Manganese Peroxidase (E.C. 1.11.1.13) Assay
Manganese peroxidase assay using phenol red was performed in the study. Five mL of reaction mixture was prepared with 1.0 mL sodium succinate buffer (50 mM, pH 4.5), 1.0 mL sodium Lactate (50 mM, pH 5.0), 0.4 mL manganese sulphate (0.1 mM), 0.7 mL phenol Red (0.1 mM), 0.4 mL H2O2 (50µM), gelatin 1 mg/mL and 0.5 mL of the enzyme extract. The reaction conducted at 30˚C was commenced by adding H2O2. One mL of the reaction mixture was added with 40µL of 5 N NaOH and absorbance was read at 610 nm. The enzyme concentration was defined in such a way that one unit of the enzyme is equivalent to an absorbance increase of 0.1units /min/ml.23
Laccase (E.C. 1.10.3.2) Assay
Laccase activity was checked by ABTS method where 2, 2′-azino-bis (3-ethylbenzothiazoline-6-sulfonic acid) diammonium salt (ABTS) acts as substrate and the absorbance were measured at 420 nm. The reaction mixture used in the method contained 100 mM sodium acetate buffer, pH 5, 0.5 mM ABTS and 400 μl enzyme supernatant. An extinction coefficient of 36,000 M-1cm-1 for ABTS was used for calculating the enzymeactivity. One unit of enzyme activity was defined as the amount of enzyme oxidizing 1 μmol of substrate per minute.24-26
Statistical Analyses
Two-way ANOVA was performed using IBM SPSS v 20 (IBM Corporation, Armonk, NY) to analyze the significant difference (Tukey test, LSD; P< 0.05) in laccase, lignin peroxidase and manganese peroxidase assays results among the samples (P. chengduensis, K. oryzendophytica and Consortium) tested at different hours (24, 48, 72 and 96).
Termite species differ in their rudimentary biology and ecology, including colony size, feeding, nesting, swarming, and reproductive behavior. Identification of termite species is a thought-provoking chore because of the vagueness in their morphological characteristics, the difficulty to distinguish morphologically distinct castes such as soldiers and alates, topographical changes, and the absence of overall methodical research for the identification of termite genera.27 The identification based on soldier morphology is considered the finest route so far compared to alates owing to seasonal existences that prevent concurrent collections and portrayal of the castes.28,29 Based on the morphological features observed under the stereomicroscope, the termites were identified as odontotermes obesus (Rambur) (Figure 1).
Figure 1. Odontotermes obesus under the stereomicroscope. A) Morphology of Thorax and abdomen. B) Head region and associated parts. C) Depiction of whole termite.
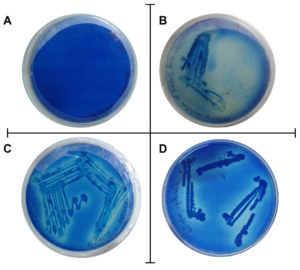
Distinct bacterial colonies with dye decolorization were observed on plates with 10-6 dilution (Figure 2). Decolorisation of methylene blue was used as a tool to identify and pick out the microorganism having lignolytic potential. In dye decolorization, the azo linkage is split either by reduction, the reaction used by some bacterial consortia under anaerobic conditions, or by oxidation, the reaction used by lignolytic white-rot fungi. Researchers found that lignolytic enzymes such as lignin peroxidase, manganese peroxidase, and laccase were responsible for dye decolorization.30 Previous scientific reports state that several bacteria are capable of dye decolorization, either in consortia or in pure cultures.31
Figure 2. Petri dish tests of oxidation of methylene blue dye. A) Control plate, B) Plates showing the oxidation of methylene blue dye by the streaked isolate, C) Isolate 1(T1), D) Isolate 2 (S).
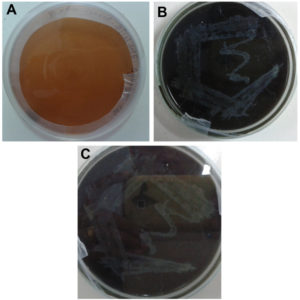
All the isolates grown in a medium containing tannic acid showed color change which indicates the presence of lignin-degrading enzymes (Figure 3). The brown color of the medium might also be due to the action of extracellular phenoloxidases which resulted in the repolymerization of degradation products.32,33
Figure 3. Plates showing Confirmatory test for lignolytic activity using tannic acid as a substitute for lignin. A) Control, B). Isolate 1(T1), C). Isolate 2 (S).
Genetic ID employing 16S rRNA has been performed for more than a decade to investigate evolutionary connections among different living beings. The comparison of rRNA sequence aid to build a universal tree of life, separating all life on earth into three equidistant domains specifically Eukarya, Bacteria, and Archaea The microbial isolates that revealed lignin-degrading ability were identified as Kosakonia oryzendophytica (GenBank Accn No. OM943747) and Pseudomonas chengduensis (GenBank Accn No. OM943748). Based on the reports by Drancourt et al.,34 when the resemblance percentage was ≥ 99% then the equivalent isolate was the same species, 97% to 99% can be specified that the analogous isolate represents the identical genus and the resemblance percentage below 97% indicates that the isolate can be declared as a new species. The BLAST alignment results show that strain T1 exhibit a similarity of 99.57% with Kosakonia oryzendophytica, strain S had a similarity of 99.93% with Pseudomonas chengduensis, respectively. The dendrogram (phylogenetic tree) was built according to the comparison of 16S rRNA sequences of the isolated strains with those extracted from the GeneBank database (HTTP:// www.ncbi.nlm.nih.gov). Out of 16S rRNA sequences from various strains, ten strains with maximum homology were compared. The dendrogram was constructed by aligning all sequences with Clustal Omega. The dendrogram represented in Figure 4 verified the linkage of the isolates T1 and S each with the other ten strains. It was found that isolate T1 was closely related to Kosakonia oryzendophytica and isolate S was closely related to Pseudomonas chengduensis respectively.
Figure 4. Dendrogram showing the 16s RNA sequencing and sequence similarities of the isolates with related genus. A) Isolate 1 (T1) K. oryzendophytica, and B) Isolate 2 (S) P. chengduensis.
Research has already proved the lignin-degrading ability of bacteria under the Enterobacter genus such as E. cancerogenus, E. ludwigii etc.35 yet no studies have been done on E. oryzendophyticus (reclassified as K. oryzendophytica36) and P. chengduensis. Microbes have developed quite a lot of enzymes for mortifying the diverse constituents of lignocellulosic material. These enzymes comprise cellulases, xylanases, and lignolytic enzymes.26 Among those, lignin peroxidase (LiP) and manganese peroxidase (MnP) have been predominantly responsible for lignin degradation.23 Liew et al. reported that manganese peroxidase produced is far more superior as compared to lignin peroxidase, suggesting that MnP might be the prevailing enzyme involved in lignin deprivation.37 A wide range of potential substrates has raised interest in the use of laccases in several industrial applications, such as delignification of pulp, bleaching of textile dye, detoxification of effluent, modification of biopolymers, and bioremediation.38,39
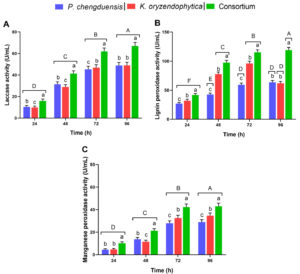
In the present study, all the bacterial isolates produced MnP, LiP, and laccase and showed a significant increase at 72 hours of incubation. The enzyme activity was found higher in the medium containing the microbial consortium than in those with the individual cultures. The consortium filtrate has MnP activity of 41.6 U/mL, LiP activity of 114.3 U/mL, and laccase activity of 61.85 U/m at 72 hours of incubation. It was found that both the enzyme activity was increased considerably until 72 hours of incubation but showed an insignificant increase at 96 hours. Hence, to attain the maximum changes in the shortest time, further analysis was performed with the optimum time of treatment as 72 hours.
The laccase, lignin peroxidase and manganese peroxidase assays results of P. chengduensis, K. oryzendophytica and Consortium were compared among the samples and the performance of individual sample at different hours (Figure 5). Overall, the P. chengduensis and K. oryzendophytica combined consortium results were highly significant within the samples and the individual sample activity at different hours (Figure 5). The individual treatments of P. chengduensis and K. oryzendophytica laccase and manganese peroxidase activities were increasing and highly significant at different hours (Figure 5a and c). On the other hand, lignin peroxidase activity of P. chengduensis and did not show a significant increase after 72 hours (Figure 5b). Conversely, K. oryzendophytica’s lignin peroxidase activity was significantly reduced after 72 hours (Figure 5b).
Figure 5. Laccase, lignin peroxidase and manganese peroxidase assays results of P. chengduensis, K. oryzendophytica and Consortium were tested at different hours. A) Laccase, B) Lignin peroxidase, and C) Manganese peroxidase activity (U/mL). Six replications (Mean±SE) were maintained for each activity. Vertical bars under different small letters were statistically significant, and the same letters indicate statistically insignificant (Tukey test, LSD; P<0.05) among the samples (P. chengduensis, K. oryzendophytica and Consortium). Different capital letters on the top of the vertical bar indicate that each sample tested at different hours (24, 48, 72 and 96) was statistically significant (Tukey test, LSD; P<0.05), same letters statistically insignificant.
Lignolytic enzymes have possible applications in various sectors such as chemical, food, fuel, paper, agricultural, textile, and other industries. Natural fibers are made of cellulose, lignin, and other constituents, of which lignin is responsible for the rigidity and hydrophobicity which in turn restricts the use of those tough fibers in various sectors of textile industries. The prominence of lignolytic bacteria has augmented since lignin-degrading bacteria have an eclectic forbearance for temperature changes, pH differences, and oxygen curb compared to fungi. The present study explored the incredible lignin-degrading capability of termites wherein the gut microflora are the real lignin biodegraders which helps the termites efficiently utilize the lignocellulosic matter. The bacteria isolated from the termite gut identified as K. oryzendophytica and P. chengduensis are found to have a strong potential for lignin degradation. Further investigations regarding the optimization of growth conditions of the bacteria might improve the lignolytic activity of the isolated strains. Thus the finding will open its path to a superior substitute for the fundamental chemical practices which benefits the future textile trade to be more eco-friendly
ACKNOWLEDGMENTS
The authors greatly acknowledge the support provided by the University Grants Commission (UGC), New Delhi, India, and the Kerala State Council for Science Technology and Environment (KSCSTE), an autonomous body of the Government of Kerala. The authors also thank Shri. D. Kuppuramu, Chairman, Coir Board, and Director, Central Coir Research Institute, India, for extending the laboratory facilities of CCRI. The authors also acknowledge the help and facilities extended by the School of Environmental Studies, Cochin University of Science and Technology, Cochin, India. We express our sincere gratitude to Dr. Sivaprasath Prabu, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing, China and Ms. Anushya AV, Research Scholar, Department of Zoology, Kannur University, Kerala for the time and support provided to mount the article.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
The article does not contain any studies with human or animal participants performed by any of the authors.
- Zhou H, Guo W, Xu B, et al. Screening and identification of lignin-degrading bacteria in termite gut and the construction of LiP-expressing recombinant Lactococcus lactis. Microbial pathogenesis. 2017;112:63-69.
Crossref - Jawaid MHPS, Khalil HPSA. Cellulosic/synthetic fibre reinforced polymer hybrid composites: A review. Carbohydr Polym. 2011:86(1):1-18.
Crossref - Rajan R, Ajith S, Joshy A, Anita R. Bleaching and softening of coir fiber using averrhoa bilimbi extract. J Nat Fibers. 2020:1-8.
Crossref - Kirk TK, Cullen D. Enzymology and molecular genetics of wood degradation by white-rot fungi. Environmentally friendly technologies for the pulp and paper industry. Wiley, New York. 1998:273-307.
- Zhou H, Guo W, Xu B, Teng Z, Tao D, Lou Y, Gao Y. Screening and identification of lignin-degrading bacteria in termite gut and the construction of LiP-expressing recombinant Lactococcus lactis. Microbial pathogenesis. 2017;112:63-9.
Crossref - Kudo T. Termite-microbe symbiotic system and its efficient degradation of lignocellulose. Biosci Biotechnol Biochem. 2009;73(12):2561-2567.
Crossref - Ohkuma M. Termite symbiotic systems: efficient bio-recycling of lignocellulose. Appl Microbiol Biotechnol. 2003;61(1):1-9.
Crossref - Yamada A, Inoue T, Wiwatwitaya D, et al. Carbon mineralization by termites in tropical forests, with emphasis on fungus combs. Eco Res. 2005;20(4):453-460.
Crossref - Ni J, Tokuda G. Lignocellulose-degrading enzymes from termites and their symbiotic microbiota. Biotechnol Adv. 2013;31(6):838-850.
Crossref - Hongoh Y. Toward the functional analysis of uncultivable, symbiotic microorganisms in the termite gut. Cell Mol Life Sci. 2011;68(8):1311-1325.
Crossref - Kohler T, Dietrich C, Scheffrahn RH, Brune A. High-resolution analysis of gut environment and bacterial microbiota reveals functional compartmentation of the gut in wood-feeding higher termites (Nasutitermes spp.). Appl Environ Microbiol. 2012;78(13):4691-4701.
Crossref - Schafer A, Konard R, Kuhnigk T, et al. Hemicellulose-degrading bacteria and yeast in the termite gut. J Appl Bacteriol. 1996:80(5):471-478.
Crossref - Borji M, S Rahimi, G Ghorbani, Yoosefi JV, Fazaeli H. Isolation and identification of some bacteria from termites gut capable in degrading straw lignin and polysaccharides. J Facul Vet Med Univ. Tehran. 2003:58:249-256.
- Wenzel M, I Schonig, M Berchtold, Kamefer P, Konig H. Aerobic and facultatively anaerobic cellulolytic bacteria from the gut of the termite Zootermopsis angusticollis. J Appl Microbiol. 2002:92(1);32-40.
Crossref - Harazono K, N Yamashita, N Shinzato, et al. Isolation and characterization of aromatics-degrading microorganisms from the gut of the lower Copiotermes formosanus. Biosci Biotechnol Biochem. 2003:67(4):889-892.
Crossref - Frohlich J, C Koustiane, P Kampfer, et al. Occurrence of rhizobia in the gut of the higher termite Nasutitermes nigriceps. J Syst Applied Microbiol. 2007:30(1):68-74.
Crossref - Ramin M, Alimon AR, Abdullah N, Panandam JM, Sijam K. Isolation and Identification of Three Species of Bacteria from the Termite Coptotermes curvignathus (Holmgren) Present in the Vicinity of University Putra Malaysia. Res J Microbiol. 2008:3(4):288-292.
Crossref - Roonwal ML, Chhotani OB. Fauna of India-Isoptera (Termites). Zoological Survey of India Publication, Calcutta. 1989;2:672.
- Chhotani OB. Fauna of India-Isoptera (Termites). Zoological Survey of India Publication, Calcutta. 1997;2:800.
- Sasikumar V, Priya V, Shankar CS, Sekar SD. Isolation and preliminary screening of lignin degrading microbes. J Academia Ind Res. 2014;3(6):291-294.
- Bandounas L, Wierckx NJ, de Winde JH, Ruijssenaars HJ. Isolation and characterization of novel bacterial strains exhibiting ligninolytic potential. BMC Biotechnol. 2011:11(1):1-11.
Crossref - Rasi RM, PU Mahalingam. Screening and Characterization of Lignin Degrading Bacteria from Decayed Sawdust. Int J Pure Appl Sci Technol. 2014:23(2):43-52.
- Arora DS, M Chander, PK Gill. Involvement of lignin peroxidase, manganese peroxidase and laccase in degradation and selective ligninolysis of wheat straw. Int Biodeterior Biodegradation. 2002:50(2);115-120.
Crossref - Mabrouk AM, Kheiralla ZH, Hamed ER, et al. Screening of some marine-derived fungal isolates for lignin degrading enzymes (LDEs) production. Agric Biol J N Am. 2010:1(4); 591-599.
- Akdogan HA, Merve C, Topuz MC, Urhan AA. Studies on decolorization of reactiveblue 19 textile dye by Coprinus plicatilis. J Environ Health Sci Eng. 2014;12(1):49.
Crossref - Rahman MM, I Jusoh, A Husaini, Ngieng SN, Seman IA. Biodegradation and ligninolytic enzymes profiles of the newly synthesized organotin (IV)-treated non durable tropical wood species. J Biochem Technol. 2014;5(3):743-750.
- Parween T, Bhandari P, Raza SK. Survey and identification of termite in some selected parts of India. Res J Life Sci Bioinform Pharm Chem Sci. 2016;2:122-135.
- Jones DT, Verkerk RH, Eggleton P. Methods for sampling termites. Insect Sampling in Forest Ecosystems. 2005:221-253.
Crossref - Norsyarizan J, Wan Nurainie WI. Morphological variation of selected species of Coptotermes (Isoptera: Rhinotermitidae) in Western Sarawak. Serangga. 2016;21:1-38.
- Dastidar MG, Simha AN, Koushik AB, et al. Bioremediation of industrial effluents by ligninolytic microbes. Int J Dev Res. 2018;08:22419-22424.
- Kumar P, Barrett DM, Delwiche MJ, Stroeve P. Methods for pretreatment of lignocellulosic biomass for efficient hydrolysis and biofuel production. Ind Eng Chem Res. 2009;48(8):3713-3729.
Crossref - Bending GD, DJ Read. Effects of the soluble polyphenol tannic acid on the activities of ericoid and ectomycorrhizal fungi. Soil Biol Biochem. 1996;28(12):1595-1602.
Crossref - Bending GD, Read DJ. Nitrogen mobilization from protein-polyphenol complex by ericoid and ectomycorrhizal fungi. Soil Biol Biochem.1996;28(12):1603-1612.
Crossref - Drancourt M, Bollet C, Carlioz A, Martelin R, Gayral J-P, Raoult D. 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J Clinical Microbiol. 2000;38(10):3623-3630.
Crossref - Ozer A, Rakici E, Bektas KI, Canakci S, Balovz AO. Isolation of lignin-degrading bacteria from different sources and testing of their ligninolytic activities. J Apitherapy Nature. 2019;2(2):30-45.
Crossref - Li CY, Zhou YL, Ji J, Gu CT. Reclassification of Enterobacter oryziphilus and Enterobacter oryzendophyticus as Kosakonia oryziphila comb. nov. and Kosakonia oryzendophytica comb. nov. Int J Syst Evol Microbiol. 2016;66(8):2780-2783.
Crossref - Liew CY, Husaini A, Hussain H, Muid S, Liew KC, Roslan HA. Lignin biodegradation and ligninolytic enzyme studies during biopulping of Acacia mangium wood chips by tropical white rot fungi. World J Microbiol Biotechnol. 2011:27(6);1457-1468.
Crossref - Xu F. Recent progress in laccase study: properties, enzimology, production and applications. In: Flickinger MC, Drew SW, editors. The encyclopedia of bioprocessing technology: fermentation, biocatalysis and bioseparation. New York: John Wiley and Sons. 1999; 1545-1554.
- Wakil SM, Adebayo-Tayo BC, Odeniyi OA, Salawu KO, Eyiolawi SA, Onilude AA. Production, characterization and purification of laccase by yeasts isolated from ligninolytic soil. J Pure Appl Microbiol. 2017;11(2):847-869.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.