ISSN: 0973-7510
E-ISSN: 2581-690X
A novel antimicrobial compounds producing strain of fungus was isolated from rhizospheric soil and identified as Aspergillus niger MTCC 12676 on the basis of morphological characters and molecular identification. Antimicrobial activity of this strain was evaluated by using different solvents against Gram positive bacteria Streptococcus mutans and Staphylococcus aureus, Gram-negative bacteria, Pseudomonas fluorescence and Escherichia coli and two yeast namely Candida tropicalis and Candida glabrata. Ethanol and ethyl acetate solvents exhibited significant antimicrobial activity against most of the tested microorganisms with more pronounced effect in case of ethyl acetate. MIC of ethanol extract ranged from 0.125-1.25 mg/ml whereas that of ethyl acetate was in the range of 0.312-0.0625 mg/ml. Antibiotic susceptibility pattern revealed that Staphylococcus aureus and Candida glabrata were resistant against the ciprofloxamine and fluconazole respectively. Combination of ciprofloxamine with different extracts (Ethanol and Ethyl acetate) exhibited synergistic effect in case of E. coli, P. fluorescence and S. mutans. Similarly, combination of the two extracts with antibiotic fluconazole had a positive synergistic effect in case of C. albicans.
Aspergillus niger, Antimicrobial activity, MIC, Synergistic effect, rhizospheric soil.
In the present world, there are many infectious diseases, re-emergent diseases and multidrug resistant bacterial diseases due to which there is continuous need of developing new antibiotics. Soil is an immense and practically unexploited source of potentially useful biological bioactive compounds. However, all biochemical chemical compounds are not well characterized. Interaction between compounds is a well-known phenomenon and substrate competition and antagonism are important factors in selection of microbial communities in any given ecological niche (Lay, 1994). Rajalakshmi and Mahesh, 2014). Development of multiple drug-resistant microbes raised the need to search for new and novel antimicrobials for treatment of human diseases. One of the major problem in health care is that emergence of antibiotic resistance bacteria Staphylococcus aureus (MRSA). MRSA is now resistant to almost all commonly used antibiotics and even resistant to last choice of antibiotics vancomycin, has begun to appear. (Denyer et al., 2004). Due to the rising incidences related to multidrug resistance amongst pathogenic microorganism, there is need to find out new antimicrobial sources (Gupta and Kumar, 2017).
Antibiotics are compounds produced by microorganisms that are able to inhibit the growth of other microorganisms (Rajalakshmi and Mahesh, 2014). In today’s world, antibiotics are the compounds produced by a microorganism, or similar substance (produced wholly or partly by chemical synthesis), whose low concentrations can inhibit the growth of other microorganisms (Barrett et al, 2005) and are considered as selective poison. Despite critical need for new antibiotics to treat drug-resistant infections and other infectious diseases, very few new antibiotics are being developed. Pathogens takes a huge variation in the time for emergence of resistance, which varies among organisms and antibiotics. When there is development of resistance in any pathogen, a new antibiotic is required, which is active against resistant one. In response to microbial resistance, the pharmaceutical industry has produced a remarkable range of antibiotics (Luzhetskyy et al, 2007). Not only is there a problem in finding new antibiotics to fight old diseases (because resistant strains of bacteria have emerged), there is also a problem to find new antibiotics to fight new diseases. Over last few years, it has become clear that the emergence of acquired resistance to antibiotics in bacteria is still a serious threat to animals and humans. So, there is urgent need to search for antibiotics which differ in their modes of action (Salyers et al, 2005).
Compounds produced from microorganisms are divided into two types: primary and secondary metabolites. Primary metabolism is governed by primary metabolites produced by microorganisms. Primary metabolism is step by step enzyme-catalyzed reactions which provide the cell with energy, synthetic intermediate products and macromolecules such as protein and nucleic acids. Products of primary metabolism are associated with cell growth and their maximum production occurs in the logarithmic phase of growth in a batch culture (Martin and Demain, 1980). In contrast to primary metabolism, secondary metabolism has no apparent function in the organism. The organism continues to exist if secondary metabolism is blocked. Many type of secondary metabolites are produced which inhibits the growth of the surrounding strains. These secondary metabolites, acts as antibiotics to maintain their predominance in the surrounding microbial population.
Several compounds with antibiotic activity have been isolated from microorganisms till date, but only a few of them are clinically useful (Morgan et al, 2005). The reason for this is because only compounds with selective toxicity can be used clinically they must be highly effective against a microorganism but have minimal toxicity to human cells. The discovery and development of new effective antimicrobial compounds with new mechanisms of action is still required for infectious disease research programs (Thomashow et al, 2008). Rhizosphere soil is mostly viewed from the perspective of how beneficial microorganisms can inhibit the growth and activity of various pathogenic microorganisms (Richardson et al, 2009). In view of the above facts, the present study reports an attempt for the isolation of potent soil fungal strain possessing antimicrobial activity against the selected human pathogenic microorganisms.
Sample collection
Soil samples for the present study were collected from different regions of Delhi, Haryana, Punjab, Himachal Pradesh and Jammu and Kashmir. During this process, wild and cultivated plants were dug out carefully and soil adhering to roots was collected removing 1-2 inches of top soil with sterilized spatula. In addition to the rhizosphere soil, sample was also collected from various other sites like bank of river, crop fields, farmhouse backyards, household wastes. A total of 150 soil samples were collected in sterile polyethylene bags and were labeled with date and site of collection. The soil samples were processed immediately for determining moisture content and pH.
Isolation of microorganisms from soil
Serial dilution agar plate method was used for the isolation of fungi from the soil samples (Cappucino and Sherman, 1996 and Aneja, 2003) through use of streptopenicillin supplemented potato dextrose agar. One gram of dry soil was suspended in 9 ml sterilized distilled water blank No. 1(stock suspension) and vortex for 10-15 minutes to obtain uniform suspension of microorganisms. 1.0 ml of suspension was transferred, while in motion, from the stock suspension (No.1), into sterile water blank number 2 with sterile pipette under aseptic conditions to make 1: 10 (10-2) dilution and vortex it well for five minutes. Further dilutions from 10-3 to 10-7 were made by pipetting 1.0 ml suspension into sterile water blanks numbered 3, 4, 5, 6 and 7 from water blanks labeled 2, 3, 4, 5 and 6 respectively. Finally 1.0 ml aliquots of the suspension of final five dilutions i.e. 10-3 to 10-7 were added to labeled and sterilized Petri plates. Approximately, 25 ml cooled (45°C) molten PDA (with streptopenicillin) was added to each Petri plate and mixed gently by rotation. After solidification of the agar media, the inoculated PDA plates were incubated in an inverted position at 25° C for 6 days. The fungi exhibiting antagonistic activity in plate with others was taken as promising isolate and cultured for further studies.
Test microorganisms
The test microorganisms used in the present study were procured from Microbial Type Culture Collection (MTCC), Institute of Microbial Technology (IMTECH), Chandigarh, India, which included Gram positive bacteria, Streptococcus mutans (MTCC Nos.497) Staphylococcus aureus (MTCC No.7443), Gram-negative bacteria, Pseudomonas fluorescence (MTCC No.1748) Pseudomonas aeruginosa (MTCC NO.1688) and Escherichia coli (MTCC No.40) and yeast namely Candida tropicalis (MTCC No. 3421),Candida glabrata (MTCC NO.3814) and Candida albicans (MTCC NO.227). The slants of modified nutrient agar were used for maintaining S. mutans strains, nutrient agar for P. flurescence, S. aureus and E. coli and malt extract agar for C. albicans strains and C. glabrata. All the slants were kept at 4 °C in the refrigerator for further studies.
Preparation of microbial inoculum
McFarland standard has been used to standardize the approximate number of microorganisms (bacteria and yeasts) in a liquid suspension by comparing turbidity of test organism suspension with McFarland standard. It is a mixture of two chemical constituents {9.95 ml barium chloride (1.175%) + 0.05 ml sulphuric acid (1.0%)}. The reaction between the two chemicals results in production of a fine precipitate of barium sulphate. When shaken well, the turbidity of 0.5 McFarland standard is visually comparable to a bacterial suspension of approximately 1.5 x 108 cells/ml. The inoculum of different test pathogens was adjusted according to above prepared 0.5 McFarland standard. The McFarland tube was stored at 4-5 °C and was prepared afresh after every 3 to 4 months (Andrews, 2001).
Preparation of crude extract
The fungal crude extract was obtained by inoculating a disc of fungal isolate in potato dextrose broth and incubating for 10-11 days at 27- 29°C in a BOD incubator. After the incubation, the biomass was filtered through Whatman filter paper No. 1. Bioactive compounds present in the filtrate were extracted by solvent extraction using ethyl acetate as a solvent. Equal volume (1:1) of culture filtrate and ethyl acetate were taken in separating funnel of 1000 ml and agitated for an hour. The mixture was allowed to stand on tripod stand till organic and aqueous layer separated out as upper and lower phase. The solvent layer was evaporated to dryness at 40 °C to get concentrated crude extract. The dried crude extract was scratched with the help of spatula and dissolved in DMSO and was used for testing of antimicrobial activity against various test pathogens.
Evaluation of antimicrobial activity
The antimicrobial activity of purified fungal isolates was determined by using agar well plate method. 100 µl of each of the test bacterial culture standardized at 10 cell/ml from 24 hour old incubated broth culture was aseptically transferred and spread over solidified Muller Hinton agar plates. Wells were dug on the seeded plate with the help of sterile cork borer (4 mm). Each well was filled with 100 µl of crude fungal extracts. The plates were incubated at 37°C for 24 to 48 hours. Antimicrobial activity of fungal isolates against test pathogens was determined by measuring the zone of growth inhibition around the fungal colonies (Corrado and Rodrigues, 2004).
Determination of minimum inhibitory concentration (MIC)
The minimum inhibitory concentration (MIC) is the lowest concentration of the antimicrobial agent that will inhibit the visible growth of a microorganism after overnight incubation (Andrews, 2001). MICs were determined following the method as described by Hirasawa et.al. (1999) with slight modifications. Different concentrations were prepared using DMSO (64, 32, 16, 8, 4, 2, 1, 0.5, 0.25, 0.125, and 0.0625mg/ml) and agar well diffusion method was carried out. The test were carried out in triplicate and the mean were recorded.
Determination of antimicrobial susceptibility and synergistic activity of promising fungal isolate with antibiotics
Antibacterial activity was measured using agar well diffusion method according to the National Committee for Clinical Laboratory Standard. Petri plates containing approximately 25-30ml of sterilized Mueller Hinton agar medium were inoculated using a cotton swab with a 4-6h old culture of the bacterial strains. Wells (6 mm diameter) were punched in the agar with sterilized cork and filled with 30 µl antibiotics and in case of synergistic effect, 30 µl of each fungal isolate was also added into the well. The plates were incubated at 37°C for 18-24 hours. The antibacterial activity was assessed by measuring the inhibition zone diameter (mm) around the well. The average of three replicates for each crude fungal metabolite, antibiotic and combination was calculated. Synergism effect was considered when combinations exhibited enlargement of combined inhibition zone size by 0.5 mm.
Identification of potential fungal isolate
The fungal strain that exhibited highest antimicrobial activity was selected for subsequent studies and was identified on the basis of its morphological, physiological, biochemical characteristics and rDNA gene sequence of ITS region. Genomic DNA was isolated and rDNA of ITS region was amplified and the PCR product was bidirectionally sequenced using forward (ITS4) and reverse (ITS5) primers The rDNA gene sequence of ITS region was used to carry out BLAST alignment search tool of NCBI Genbank database. Based on maximum identity score first fifteen sequences were selected and aligned using multiple alignment software program Clustal W. Distance matrix was generated using RDP database and the phylogenetic tree was constructed using MEGA 5.
Selection of Promising fungal isolates
Eight fungal isolates: A21 (MTCC 12676), A23, A63, A72, A129 (Aspergillus spp.), A56 (Penicillium spp.) and A103, A113 (Cladosporiun sp.) were found to be effective against test pathogens. On the basis of maximum diameter of zone of growth inhibition against test pathogens using agar well diffusion assay obtained from screening studies, the fungal isolate A21 proved most promising and was selected for further study.
Identification of fungal isolate
Identification of the fungal isolate A21 was done on the basis of colony characteristics (colony growth, colour and production of exudate) and sporulating structures (conidiogenous cells, vesicle, conidial head and conidia) by following manual of Kirk et al. (2001). Morphological characterization suggests that it is a fungal strain belonging to Aspergillus genera. For molecular analysis, the rDNA of ITS region was amplified and the PCR product was bidirectionally sequenced using forward (ITS4) and reverse (ITS5) primers. Based on BLAST search analysis the fungus isolate A21 was identified as Aspergillus niger. Based on maximum identity score first 90 sequences were selected and aligned using multiple sequence alignment software program clustal W. Distance matrix was generated using RDP database and the phylogenetic tree was constructed using muscle (Fig. 1). A living culture of the isolate is deposited in Microbial Type Culture Collection (MTCC), Institute of Microbial Technology (IMTECH) Chandigarh, India (Accession No.12676).
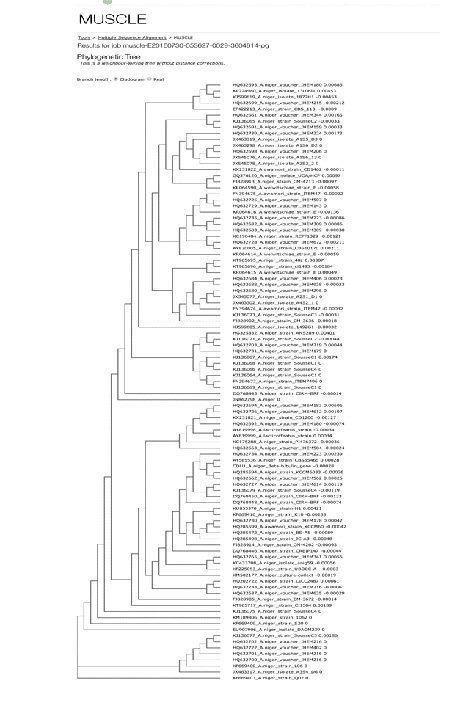
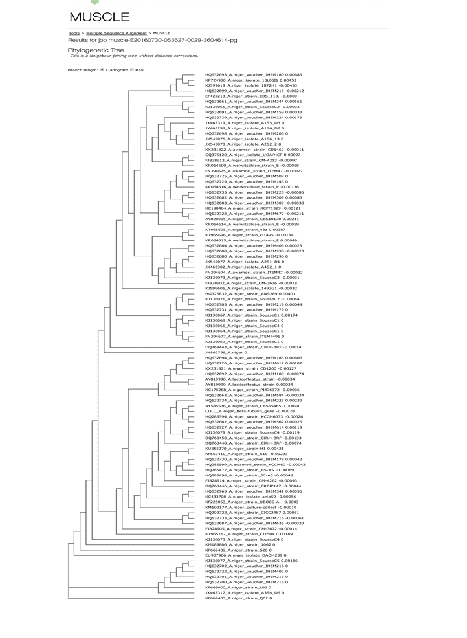 Fig. 1. Phylogenetic tree of Aspergillus niger MTCC 12676 Determination of antimicrobial activity
Fig. 1. Phylogenetic tree of Aspergillus niger MTCC 12676 Determination of antimicrobial activityDetermination of antimicrobial activity
In the present study, crude extracts of Aspergillus niger MTCC 12676 were prepared in different solvents (Methanol, petroleum ether, ethanol, n-butanol and ethyl acetate) and analyzed for their antimicrobial activity determined against the test microbial pathogens. The results presented in Table 1 shows that antimicrobial activity of the fungal strain A. niger MTCC 12676 was obtained only in case of ethanol and ethyl acetate solvents. These extracts exhibited significant antimicrobial activity against most of the tested microorganisms. In ethanolic extracts, the zone of growth inhibition ranged from 14-24 mm whereas the ethyl acetate extracts resulted in zones of 16-36 mm. Among the tested pathogens, Candida albicans was found to be most sensitive yielding 24 mm and 36 mm diameter zones against ethanol and ethyl acetate extracts respectively. However, none of the solvent was able to exhibit the antimicrobial activity against Pseudomonas aeruginosa and Candida tropicalis. There are also previous reports suggesting the antimicrobial activity of Aspergillus sp. against the pathogenic microorganisms (Furtado et al., 2002 and Sunkar and Nachiyar, 2011).
Table (1):
Antimicrobial activity of different crude extracts of A. niger MTCC 12676 against various pathogenic microorganisms.
| Solvent | Diameter of zone of inhibition (mm) | |||||||
|---|---|---|---|---|---|---|---|---|
| Gram-negative | Gram-positive | Yeast | ||||||
| Ec | Pa | Pf | Sa | Sm | Ct | Ca | Cg | |
| Methanol | – | – | – | – | – | – | – | – |
| Petroleum Ether | – | – | – | – | – | – | – | – |
| Ethanol | 21±0.23 | – | 18±0.1 | 19±0.23 | 20±0.42 | – | 24±0.13 | 14±0.21 |
| n-Butanol | – | – | – | – | – | – | – | – |
| Ethyl Acetate | 29±0.47 | – | 24±0.31 | 21±0.21 | 26±0.21 | – | 36±0.37 | 16±0.23 |
Ec: Escherichia coli, Pa: Pseudomonas aeruginosa, Pf: Pseudomonas florocesence, Sa: Staphylococcus aureus, Sm: Streptococcus mutans, Ct: Candida tropicalis Cs: Candida albicans, Cg: Candida glabrata
*Mean diameter± Standard deviation
Determination of minimum inhibitory concentration (MIC)
The minimum inhibitory concentration of ethanolic extract of A. niger MTCC 12676 and of ethyl acetate extracts are shown in Table 2. MIC of ethanol extract was 0.125 mg/ml for Escherichia coli, Staphylococcus aureus and 1.25mg/ml for Pseudomonas florocesence, Streptococcus mutans and Candida albicans. The lowest MIC was observed in case of Escherichia coli (.125mg/ml). On the other hand, MIC of ethyl acetate extract was 0.312mg/ml for Escherichia coli, Pseudomonas florocesence, Candida albicans and 0.0625mg/ml for Staphylococcus aureus and Streptococcus mutans. Candida glabrata had highest MIC of 2.5mg/ml among all the tested microorganisms in both the solvents. The present results are in accordance to Geetanjali et al. (2016) who reported fungal antimicrobial activity against most of the microorganisms used in the present study with a closely matched MIC values.
Table (2):
Minimum inhibitory concentration (MIC) of crude extracts of A. niger MTCC 12676 prepared in different solvents against the test pathogens.
| Solvent | Concentration (mg/ml) | |||||||
|---|---|---|---|---|---|---|---|---|
| Gram-negative | Gram-positive | Yeast | ||||||
| Ec | Pa | Pf | Sa | Sm | Ct | Ca | Cg | |
| Methanol | – | – | – | – | – | – | – | – |
| Petroleum Ether | – | – | – | – | – | – | – | – |
| Ethanol | 0.125 | – | 1.25 | 0.125 | 1.25 | – | 1.25 | 2.5 |
| n-Butanol | – | – | – | – | – | – | – | – |
| Ethyl Acetate | 0.312 | – | 0.312 | 0.0625 | 0.0625 | – | 0.312 | 2.5 |
Ec: Escherichia coli, Pa: Pseudomonas aeruginosa, Pf: Pseudomonas florocesence, Sa: Staphylococcus aureus, Sm: Streptococcus mutans, Ct: Candida tropicalis Cs: Candida albicans, Cg: Candida glabrata
*Mean diameter± Standard deviation
Determination of antimicrobial susceptible pattern of tested microorganisms
Table 3 reveals the antibiotic susceptibility pattern of test pathogens. In case of gram–positive bacteria, Pseudomonas aeruginosa showed the maximum susceptibility to antibiotic ciprofloxacine producing inhibition zones of 32 mm, whereas Streptococcus mutans had zone of 28 mm. On the other hand, Gram – negative bacteria, exhibited zone of 30 mm and 26 mm for Escherichia coli and Pseudomonas fluorescence respectively against the same antibiotic. However, Staphylococcus aureus was found to be resistant against ciprofloxamine. Similarly, he fungi Candida tropicalis (26 mm) and Candida albicans (29 mm) also exhibited susceptibility pattern against antibiotic fluconazole except Candida glabrata.
Table (3):
The antibiotic susceptibility pattern of test microorganisms.
STRAINS USED |
CIPROFLOXACINE |
FLUCONAZOLE |
DIAMETER OF ZONE OF INHIBITION (mm)*** |
|---|---|---|---|
Escherichia coli |
+ |
– |
30±0.33 |
Pseudomonas aeruginosa |
+ |
– |
32±0.67 |
Pseudomonas fluorescence |
+ |
– |
26±0.37 |
Staphylococcus aureus* |
+ |
– |
NA |
Streptococcus mutans |
+ |
– |
28±0.45 |
Candida tropicalis |
+ |
26±0.23 |
|
Candida albicans |
+ |
29±0.13 |
|
Candida glabrata.** |
+ |
NA |
*Staphylococcus aureus: resistant to Ciprofloxacin
**Candida glabrata: resistant to Fluconazole
***Mean Diameter ± Standard Deviation
NA: No activity
Synergistic activity of promising fungal isolate with antibiotics
The synergistic activities of ethanol and ethyl acetate extracts of A. niger MTCC 12676 with commercial antibiotics were determined to observe changes in their antimicrobial efficacy (Table 4). When both the extracts were combined with ciprofloxamine, no synergistic effect was observed in case of Gram-negative bacterium P. aeruginosa and Gram-positive S. aureus. However, this combination of ciprofloxamine with different extracts (Ethanol and Ethyl acetate) exhibited synergistic effect through significant inhibition of the growth of Gram-negative bacteria E. coli, P. fluorescence and Gram- positive bacterium S. mutans as indicated by significant increase in the diameter of the zone of inhibition. Similarly, a positive synergistic effect was observed in case of C. albicans, where zone of inhibition increased significantly in response to the combination of the two extracts with antibiotic fluconazole. However, no effect was observed in case of C. tropicalis and C. glabrata.
Table (4):
Synergistic effect of different solvent extracts of A. niger MTCC 12676 with selected antibiotics.
| STRAINS USED | CIPROFLOXACINE | FLUCONAZOLE | CRUDE EXTRACT | DIAMETER OF ZONE OF INHIBITION (mm)*** | |
|---|---|---|---|---|---|
| ET | EA | ||||
| Escherichia coli | + | – | + | 32±0.21 | 34±1.09 |
| Pseudomonas aeruginosa | + | – | + | 32±0.21 | 32±0.21 |
| Pseudomonas fluorescence | + | – | + | 27±0.67 | 28±0.76 |
| Staphylococcus aureus* | + | – | + | 18±0.78 | 22±0.33 |
| Streptococcus mutans | + | – | + | 30±0.23 | 32±0.21 |
| Candida tropicalis | – | + | + | 26±.43 | 26±.43 |
| Candida albicans | – | _ | + | 38±1.09 | 38±1.09 |
| Candida glabrata.** | – | + | + | 13±0.65 | 16±0.76 |
Abbreviations: ET: Ethanol, EA:Ethyl acetate
*Mean Diameter ± Standard Deviation
A number of Aspergillus species have been studied to be the potent source of antimicrobial compounds, but a lot more still remained to be explored. In the present study, a novel strain of Aspergillus niger MTCC 12676 was isolated from the soil and investigated for the antimicrobial activity against some disease causing microorganisms. The results obtained indicates that the crude extract of this fungi was very effective against the growth of these microorganism and could represent a possible source of antimicrobial compounds against the tested microorganisms. It was also revealed that fungi also inhibited the growth of Staphylococcus aureus and Candida glabrata both of which exhibited resistance against the commercial antibiotics ciprofloxacine and fluconazole, respectively. Further studies are required for the isolation and characterization of the antimicrobial compounds which could be used for pharmaceutical applications for treatment and control of infectious disease causing bacteria and fungus.
ACKNOWLEDGMENTS
The authors thank Chairperson, Department of Biotechnology, Chaudhary Devi Lal University, Sirsa, Haryana, India for providing necessary laboratory facilities to carry out this work.
- Lay, W.B. Microbes analysis in laboratory. Raja Grafindo Persada, Jakarta, Indonesia, 1994.
- Rajalakshmi, S., Mahesh, N. Production and characterization of bioactive metabolites isolated from Aspergillus terreus in rhizosphere soil of medicinal plants. Int. J. Curr. Microbiol. Appl Sci., 2014; 3(6): 784-798.
- Denyer, S.P., Hodges, N.A., German, S.P. Hugo and Russell’s Pharmaceutical Microbiology, 7th edn. Blackwell Science, India, 2004.
- Gupta, D., Kumar, M. Evaluation of in vitro antimicrobial potential and GC–MS analysis of Camellia sinensis and Terminalia arjuna. Biotechnol. Reports., 2017; 13: 19–25
- Barrett, J.F. MRSA – what is it, and how do we deal with the problem? Expert Opin. Ther. Targets., 2005; 9: 253 – 265.
- Luzhetskyy, A., Pelzer, S., Bechthold, A. The future of natural products as a source of new antibiotics. Curr. Opin. Investig. Drugs., 2007; 8: 608-613.
- Salyers, A.A., Whitt, D.D. Revenge of the microbes: how bacterial resistance is undermining the antibiotic miracle. ASM Press, Washington D.C., 2005.
- Martin, J.F., Demain, A.L. Control of antibiotic synthesis. Microbiol. Rev., 1980; 44: 230 – 231.
- Morgan, J.A.W., Bending, G.D., White, P.J. Biological costs and benefits to plant–microbe interactions in the rhizosphere. J. Exp. Bot., 2005; 56: 1729–1739.
- Thomashow, L.S., Bonsall, R.F., David, M. Secondary metabolites in soil ecology. Springer. Berlin Heidelberg, 2008.
- Richardson, A.E., Barea, J.M., McNeill, A.M., Prigent-Combaret, C. Acquisition of phosphorus and nitrogen in the rhizosphere and plant growth promotion by microorganisms. Plant Soil, 2009; 321: 305–339.
- Cappuccino, J.G., Sherman, N. Microbiology – A Laboratory manual, The Benjamin/Cummings Publishing Co., Inc., Menlo Park, California, 1996.
- Aneja, K.R. Experiments in microbiology and plant pathology. New Age International Pvt. Ltd., New Delhi, 2003.
- Andrews, J.M. Determination of minimum inhibitory concentration. J. Antimicrob. Chemother., 2001; 48: 5-16.
- Corrado, M., Rodrigues, K.F. Antimicrobial evaluation of fungal extracts produced by endophytic strains of Phomopsis sp. J. Basic Microbiol., 2004; 44: 157-160.
- Hirasawa, M., Shouji, N., Neta, T., Fukushima, K., Takada, K. Three kinds of antibacterial substances from Lentinus edodes (Berk.) Sing. (Shiitake, an edible mushroom). Int. J. Antimicrob. Agents, 1999; 11(2):151-7.
- Furtado, N.A.J.C., Said, S., Ito, I.Y., Bastos, J.K. The antimicrobial activity of Aspergillus fumigatus is enhanced by a pool of bacteria. Microbiol. Res., 2002; 157(3): 207-211.
- Sunkar, S., Nachiyar, C.V. Isolation and characterization of antimicrobial compounds produced by endophytic fungus Aspergillus Sp. isolated from Writhtia tintorica. J. Pharm. Res., 2011; 4(4): 1136-1137
- Geetanjali, Jain, P., Kumar, T. Evaluation of antimicrobial potential of rhizospheric soil fungi isolated from Tinospora cordifolia, Mentha arvensis and Ocimum tenuifloram medicainal plants. Int. Res. J. Pharm., 2016; 7(9).
© The Author(s) 2017. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.


