ISSN: 0973-7510
E-ISSN: 2581-690X
This study isolated and identified Bacillus amyloliquefaciens B1 from Carcinoscorpius rotundicauda by carrying out the 16S rRNA sequence analysis, reconstructing the phylogenetic tree based on the Environment for Tree Exploration (ETE3) v3.1.1 belonging to the GenomeNet. By an indirect competitive enzyme-labeled immunoassay, B1 could produce tetrodotoxin (TTX) in MRS was more highly than LB media. After purification, TTX producing ability in B1 could be detected in ELISA assay, high performance liquid chromatography (HPLC). The gel permeation chromatography and gas chromatography were applied to determine the molecular weight of EPS and the concentration of glucose in EPS. The results indicated the highest molecular weight of exopolysaccharides (EPS) estimated 1.33 × 106 Da consisted of glucose (150.09 µg/g). TTX yield was proportional to EPS production in the bacterium. The antimicrobial activities of EPS were determined by agar well diffusion method. Diameter of inhibition zone (mm) of Bacillus amyloliquefaciens EPS on the test microorganisms. The EPS could inhibit against Staphylococcus aureus ATCC 25923, Pseudomonas aeruginosa ATCC 27853, Escherichia coli ATCC 25922, Vibrio parahaemolyticus ATCC 17802 and Micrococcus luteus ATCC 10240. In silico prediction, TTX might interact with Bacillus amyloliquefaciens via the extracellular domain of noncanonic ABC-type transporter from gram positive bacteria. TTX might also interact with peptidase S54, mistic, metal binding protein of Bacillus subtilis and tryptophan-rich sensory protein of Bacillus cereus. This study provides the understanding of TTX producing Bacillus amyloliquefaciens B1 isolated from Carcinoscorpius rotundicauda.
Carcinoscorpius rotundicauda, Bacillus amyloliquefaciens B1, Tetrodotoxin, Exopolysaccharides, Characterization
Carcinoscorpius rotundicauda belongs to horseshoe crab found commonly in Thailand, Malaysia, Vietnam, Indonesia, China.1 They have a hard exoskeleton and a long, spiny tail, which are characteristic features of the horseshoe crab family. Carcinoscorpius rotundicauda is typically brown to gray in color and can grow up to 20 cm in length.2 Carcinoscorpius rotundicauda produces tetrodotoxin (TTX) and TTX analogues2 that causes toxicity for human although TTX is being exploited in health care.3,4
TTX is a potent neurotoxin that is found in various species of marine and freshwater animals, including pufferfish,5 xanthid crab,6 blue-ringed octopus,7 flatworm,8 newts,9 gastropods.10 This toxin is responsible for the symptoms of TTX poisoning, which can range from mild symptoms such as tingling of the lips and fingers to severe symptoms such as paralysis, respiratory failure, and death. The exact mechanism of action of tetrodotoxin is not completely understood, but it is known to block the sodium channels in the nerve cell membrane, leading to paralysis and death.11,12 The toxin is heat-stable, meaning that it cannot be destroyed by cooking, and it is also resistant to most forms of chemical degradation. This makes it a particularly dangerous toxin, as it can persist in contaminated food for long periods of time. In addition to its toxicity to humans, TTX has been studied for its potential medical applications. It is being studied as a potential treatment for chronic pain.13-15
Tetrodotoxin (TTX) is studied on the production by several microorganisms, mostly bacteria.16-20 The exact mechanism of TTX production by these microorganisms is not yet fully understood, but several studies have attempted to explain. In bacteria, TTX production was associated with the plasmid copy numbers in Aeromonas21 or suspected role of non-ribosomal peptide synthetic (NRPS) and polyketide synthase (PKS) in Vibrio alginolyticus.22 TTX is believed to be produced as a secondary metabolite in response to stress factors such as nutrient limitation, high light intensity, and exposure to pollutants. Overall, the mechanisms of TTX production by microorganisms are complex and involve the interaction of multiple genetic and environmental factors. Microorganisms which are symbiotic with host are studied on the products as host.23,24 In recent years, endophytic microorganisms have gained significant attention as potential sources of novel bioproducts and biotechnological tools. For example, endophytic fungi have been found to produce secondary metabolites with medicinal properties, such as antibiotics, anticancer agents, and anti-inflammatory agents. In bacteria, exopolysaccharides are complex sugars and often found in their extracellular environment.25 EPS production is a common mechanism used by bacteria to form biofilms, which provide them with protection, communication, and enhanced resistance to environmental stressors. Some species of bacteria can produce EPS that have therapeutic properties and can be used as a source of food, feed, or biodegradable materials. However, EPS can also play a role in bacterial pathogenesis and antibiotic resistance by shielding bacteria from the host immune response and from the action of antibiotics. Some bacteria, such as Pseudomonas and Vibrio species, produce TTX, which also produces EPS. EPS involved TTX toxicity26 that might provide a protective barrier around the bacteria and help them colonize specific environments. It is important to note that further research is needed to fully understand the role of exopolysaccharides in the production and distribution of tetrodotoxin, and to determine the potential implications for human health and the environment.
From the above bases, this study isolated, identified and characterized the TTX producing microorganism to give another object to study more TTX producing sources. The study performed in-silico prediction on TTX interaction with membrane protein to estimate the reason why TTX can exist in bacteria. Besides that, EPS of isolate was also determined and found the TTX yield and EPS proportion. Antimicrobial activity was checked to explain why host was not infected with other microorganisms except the isolated microorganism.
Sample Preparation for isolation
The egg fluid from Carcinoscorpius rotundicauda collected at the seas of Vietnam was used to culture for bacterial detection. There is no specific permission required for the studies. The Carcinoscorpius rotundicauda organisms were weighed around 0.5-1 kg, kept alive before performance. The organisms were cleaned with 70% ethanol and operated to take out the fresh egg fluids. In the study, egg fluids were used to culture for bacterial isolation. The fluid (1 mL) was added and incubated directly in modified MRS and LB broth. The reasons for selecting media due to the previous study that NaCl and KH2PO4 let microorganisms grow well to produce TTX.27 Cultures were incubated for 48-72 hours in optimal conditions (aerobic and 5% CO2) at 37°C. Then, the cultures were checked for searching the microorganism on MRS and LB agar.
Isolation and Identification of bacteria
One inoculum (100 µL) of culture was carefully spread on LB and MRS agar plates and incubated at 25°C and 37°C for 2–5 days.
Nextly, the colonies appeared in the plates were picked and spread onto another MRS and LB agar plates, respectively. All the plates were observed to check the colony growth. Each colony was then subcultured in media to check TTX production by using Tetrodotoxin (TTX) ELISA Test Kit (Meizheng, Perkin Elmer, China). The expected strain was identified by morphological, biochemical and molecular characterization. The biochemical tests were done using API 50 CHB (bioMerieux, Lyon, France). For molecular study, 16S rRNA sequence was done and analyzed. The forward reverse primers were designed based on the morphology and biochemical test. The conditions for PCR were set as the previous study.19,20 The 16S rRNA sequence was compared with the homology to the almost 16S rRNA sequences using the NCBI BLAST database. The 16S rRNA sequence was also analyzed for the phylogeny by using the Environment for Tree Exploration (ETE3) v3.1.1 belonging to the GenomeNet.28,29 The 16S rRNA sequence was deposited in DDBJ (Japan).
Tetrodotoxin Extraction and Purification from Bacterial Cultures
After culturing bacteria in LB and MRS media for 2-5 days, the obtained cultures (1012 cfu/mL) were centrifuged at a suitable speed to collect supernatants which were called extracts. The extracts obtained from LB and MRS were heated at 105°C and freeze-dried for ELISA assay according to the instruction of manufacturer (Meizheng, Perkin Elmer, China). Moreover, TTX from treated extracts was mixed with acetic acid (1%) at the ratio (1:2 w/w) to perform column chromatography using charcoal as the previous performance.20 TTX was recovered from charcoal by eluting in 1% acetic acid. The eluate was evaporated, freeze-dried, and then dissolved in 1% acetic acid for qualitative and quantitative analysis. To confirm TTX in the extract, HPLC was also carried out.19,20
In-silico prediction of the interactions of TTX with some bacterial membrane proteins
The screening was carried out based on the previous method.30-32 TTX was utilized as ligand. The Protein Data Bank (PDB) and the PubChem website were used to get the three-dimensional structures of membrane proteins. This experiment included as protein targets from the PDB. These proteins were chosen for study because they were thought to involve TTX transportation and cause the gene expression for TTX production inside bacteria for further studies. Using Autodock 4.2, 5 docking experiments were conducted to evaluate the binding energy of substrates to proteins. Using Discovery Studio Visualizer, the results and interactions were then evaluated (2D, 3D).
EPS extraction
EPS was collected from bacterial cultures by using ethanol precipitation method with modification.26,33,34 Basically, the cell free supernatant of bacterium was mixed with absolute ethanol with the ratio 1:2 at 4°C for 24 hours. After that, the mixture was centrifuged at 12,000 rpm for 30 minutes at 4°C. Pellets were then collected for an antimicrobial activity test. After that, DNase, RNase, and protease were added to remove DNA, RNA, and proteins. These enzymes were then removed by heating at 100°C for 30-60 minutes. The pellets were weighted and dissolved in distilled water to get soluble EPS. Phenol-sulfuric acid method was used to estimate the total carbohydrate. For molecular weight and monomer determination, gel permeation chromatography and gas chromatography were performed for analysis.
Phenol-sulfuric acid assay for total carbohydrate detection
The amount of total carbohydrate was measured by using phenol-sulfuric acid method. This method was first described by Dubois et al.35 and later adapted to a 96-well application by Masuko et al.36 D-glucose (100 μg/mL) was used as standard for determination of carbohydrate. In each well of 96-well plate, 50 µL of EPS sample and glucose solution were added into each well where 150 µL of sulfuric acid and 30 µL of phenol 5% (w/v) were added then. After incubating for 5 min at 90°C in a water bath by floating the microplate carefully, the plate was cooled for 5 min at room temperature and measured at the wavelength of 490 nm by microplate reader. Based on the graph of standard D-glucose, the concentration of EPS samples was determined.
Determination of molecular weight of exopolysaccharides (EPS)
Gel permeation chromatography (GPC) was used to ascertain the molecular weight of EPS. Crude EPS samples were dissolved in distilled water which was used as mobile phase. The solution was put into GPC device at flow rate of 0.6 mL/min. The molecular weight of EPS was determined.26,37
Determination of the glucose monomer in EPS
The glucose and its amount in EPS were certified by gas chromatography with flame ionization detector (GC-FID) assay.38 EPS solution (20 mg/mL) was hydrolyzed at 100°C with HCl for 3 hours. The mixture was then dried to give derivative with acetic anhydride in a pyridine medium and analyzed by GC-FID. The amount of glucose was then determined.
Antimicrobial activity detection
The antimicrobial activity of EPS was evaluated by agar well diffusion method.39-41 Both unheated and heated EPS samples were used for an antimicrobial activity test. Erythromycin (512 mg/mL) and distilled water were used for positive and negative control, respectively. After incubation, the inhibition zones were noted, and scaled to obtain the zone diameter in millimeter. The pathogens including Staphylococcus aureus ATCC 25923, Pseudomonas aeruginosa ATCC 27853, Escherichia coli ATCC 25922, Vibrio parahaemolyticus ATCC 17802 and Micrococcus luteus ATCC 10240 were applied in the study.
Data analysis
All the data was analyzed by using SPSS (Statistical Package for the Social Sciences) and Microsoft Excel. All experiments were replicated, and results were performed by Mean + Standard Deviation (SD).
Identification of bacterium
After separateness, the colonies were progressed in media to check TTX producing capability by using ELISA analysis. There were 4 colonies producing TTX in MRS (Figure 1). The colony which gave the loftiest yield, calculated as 207 mg TTX in 1 L culture when cultivated in MRS medium was identified. The remaining colonies will be confirmed and optimized for TTX producing culture in the future.
The expected strain was identified by morphological and biochemical characterization. Based on the morphological observation, the isolated strain was rod-shaped and could produce spores as Bacillus. With the biochemical identification, the bacterium showed the fermentation with glycerol, L-arabinose, D-ribose, D-xylose, D-galactose, D-glucose, D-fructose, D-mannitol, N-acetyl glucosamine, arabinose, esculin, salicin, D-cellobiose, D-Trehalose, D-raffinose, and amidon, suggested the isolated strain might be Bacillus subtilis or Bacillus amyloliquefaciens according to the guidelines of the kit. For more confirmation, molecular characterization was proceeded. The accession number (LC757017) of 16S rRNA sequence was assigned by DDBJ. The 16S rRNA sequence was compared with the homology to the nearly 16S rRNA gene sequence in the NCBI BLAST database. From that, the 16S rRNA gene sequence was also aligned with some relating bacteria for the phylogeny by using the Environment for Tree Exploration (ETE3) v3.1.1 belonging to the GenomeNet. Figure 2 showed the similarities of the 16S rRNA gene sequence of isolate with the sequences of Bacillus amyloliquefaciens and others in genebank. Using the phylogeny tree analysis, the isolate was closed to Bacillus amyloliquefaciens (Figure 3). Through alignment and phylogeny results, the isolate was named Bacillus amyloliquefaciens B1. The TTX production from B1 was confirmed by HPLC. By comparing to the retention time of standard TTX (13.376 mins), the partial purified culture of isolate gave the retention time (13.330 mins) that was similar to standard TTX, suggesting the TTX actuality in the extract. It was meant that the isolated strain could produce TTX.
Figure 2. Multiple alignment. CP009748.1: Bacillus subtilis strain ATCC 13952; CP 002927.1: Bacillus amyloliquefaciens XH7; FN 597644.1: Bacillus amyloliquefaciens DSM7; CP116011.1: Bacillus amyloliquefaciens strain SRCM124317; CP021505.1: Bacillus amyloliquefaciens strain SRCM101267; CP018902.1: Bacillus amyloliquefaciens strain HK1; CP035393.1: Bacillus velezensis strain SRCM103691; CP035399.1: Bacillus velezensis strain SRCM103788; CP041693.1: Bacillus amyloliquefaciens strain H; CP035410.1: Bacillus velezensis strain SRCM103616; CP044444.1: Bacillus amyloliquefaciens strain KC41; CP054415.1: Bacillus amyloliquefaciens strain 205; CP082279.1: Bacillus amyloliquefaciens strain Bam1
Figure 3. Phylogeny analysis. CP082279.1: Bacillus amyloliquefaciens strain Bam1; FN 597644.1: Bacillus amyloliquefaciens DSM7; CP054415.1: Bacillus amyloliquefaciens strain 205; CP035399.1: Bacillus velezensis strain SRCM103788; CP116011.1: Bacillus amyloliquefaciens strain SRCM124317; CP021505.1: Bacillus amyloliquefaciens strain SRCM101267; CP009748.1: Bacillus subtilis strain ATCC 13952; CP035410.1: Bacillus velezensis strain SRCM103616; CP041693.1: Bacillus amyloliquefaciens strain H; CP035393.1: Bacillus velezensis strain SRCM103691; CP044444.1: Bacillus amyloliquefaciens strain KC41; CP 002927.1: Bacillus amyloliquefaciens XH7; CP018902.1: Bacillus amyloliquefaciens strain HK1
In-silico study for TTX interacting with Bacillus membrane proteins
From the results searched in PDB, Bacillus amyloliquefaciens had the crystal structure of YknZ (5F9Q), the extracellular domain of noncanonic ABC-type transporter from gram-positive bacteria. The residues 103-219 formed the upper subdomain in which a seven-stranded antiparallel β-sheet was observed with four α-helices. There was a loop in which the significant electron density was not seen connected β2 to β9 in this domain. The residues 58-102 and 219-256 created lower subdomain that consisted of three α-helices and a three-stranded antiparallel β-sheet. The protein could bind to TTX by van der Waals, carbon hydrogen bond and conventional hydrogen bond (Figure 4).
Figure 4. Modeling interaction of the extracellular domain of noncanonic ABC-type transporter from Bacillus amyloliquefaciens and TTX
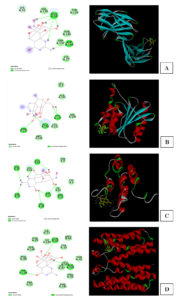
Figure 5. Docking for interaction of TTX with some membrane proteins in Bacillus species. (A) Peptidase S54 of Bacillus subtilis; (B) Mistic protein of Bacillus subtilis subsp. subtilis str. 168 (C) Tryptophan-rich sensory protein of Bacillus cereus ATCC 14579 (D) The uncharacterized domain of unknown function 1775 (DUF1775) containing YcnI, a metal binding protein of Bacillus subtilis
Moreover, to exploit more the membrane proteins which might involve the TTX interaction to bacterium, the study also checked some membrane proteins of other Bacillus species that had the identities to primary sequences of Bacillus amyloliquefaciens. These proteins were mentioned in Table 1.
Table (1):
Membrane proteins in Bacillus species having similarities to Bacillus amyloliquefaciens.
Membrane Protein |
Species |
ID |
Identities |
Positives |
Protein |
|---|---|---|---|---|---|
The N-terminal domain of rhomboid protease YqgP |
B. subtilis |
6R0J |
61/181(34%) |
61/181(34%) |
Peptidase S54 |
NMR structure of Mistic |
Bacillus subtilis subsp. subtilis str. 168 |
1YGM |
64/80(80%) |
73/80(91%) |
Mistic |
Crystal structure of BcTSPO/PK11195 complex |
Bacillus cereus ATCC 14579 |
4RYI |
99/155(64%) |
122/155(78%) |
Tryptophan-rich sensory protein |
The uncharacterized domain of unknown function 1775 (DUF1775) containing YcnI, a metal binding protein |
Bacillus subtilis |
7MEK |
110/127(87%) |
115/127(90%) |
DUF1775 domain-containing protein |
Figure 5A showed the interaction of those proteins with TTX. The domain of YcnI, called metal binding protein (7MEK) in Bacillus subtilis had the active sites including His27, Glu50, Trp137. The N-terminal domain of rhomboid protease YqgP (6R0J) of Bacillus subtilis showed the interaction as in Figure 5B. Figure 5C showed NMR structure of mistic (1YGM) of Bacillus subtilis subsp. subtilis str. 168. Mistic is a membrane-integrating protein showed the active sites which might interact with TTX. In Figure 5C, Glu110 at the C terminus was well exposed periplasmically. Cys3 at the N terminus of the protein and the centrally located Ser58, both also evidently on the extracellular side of the membrane, were nonreactive with membrane protein binding site in the right-side-out (RSO) membrane vesicles, consistent with these side chains being embedded in the membrane. Crystal structure of BcTSPO/PK11195 complex (4RYI) of Bacillus cereus ATCC 14579 had ligand binding sites as Y32, W51, F55, N87, F90, W138, A142 (Figure 5).
EPS detection
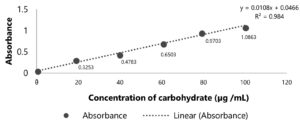
The study also characterized EPS because EPS involving the TTX toxicity involvement.26 By detecting carbohydrate using Phenol-sulfuric acid assay, the concentration of EPS was determined (Figure 6). The standard line had the equation y=0.0108x+0.0466 with R2=0.984 >0.97, which meant this standard curve was reliable and can be used for calculating the concentration of EPS. The concentration of EPS in supernatant was 89.33 + 7.53 (µg /mL).
Determination of molecular weight and monomer of EPS
The molecular weight of Bacillus amyloliquefaciens B1 exopolysaccharides (EPS) was determined by gel permeation chromatography method (GPC). The result was shown in Figure 7. The maximum molecular weight of EPS was calculated about 1.3×106 Da.
The amount of glucose in EPS was determined by gas chromatography (Figure 8). The concentration of glucose was calculated as 150.90 µg/g EPS.
Figure 9. Antimicrobial activity of EPS. (A). Micrococcus luteus. (B). Vibrio parahaemolyticus. (C). Staphylococcus aureus. (D). Escherichia coli. (E). Pseudomonas aeruginosa
Antimicrobial activities of EPS
After antimicrobial activity performance, the results of the heated and unheated samples were not significant. Therefore, the data of the heated EPS was expressed in Table 2 and Figure 9.
Each data was expressed as mean + standard deviation. The concentration of positive control was based on Table 2. The concentration of EPS was 4mg/ml of distilled water.
Table (2):
Diameter of inhibition zone (mm) of Bacillus amyloliquefaciens EPS.
| Pathogens | Inhibition zone (mm) | |
|---|---|---|
| EPS | Positive control | |
| Escherichia coli | 5.50 + 0.58 | 7.75 + 0.29 |
| Micrococcus luteus | 9.75 + 0.06 | 22.5 + 0.58 |
| Pseudomonas aeruginosa | 4.95 + 0.06 | 28.50 + 0.58 |
| Vibrio parahaemolyticus | 3.28 + 0.22 | 20.5 + 0.58 |
| Staphylococcus aureus | 14.55 + 0.29 | 26.55 + 0.29 |
In this study, we have identified the toxicity of Bacillus amyloliquefaciens B1 isolated from Carcinoscorpius rotundicauda collected in Khanh Hoa sea. It could produce TTX, contributing the speculation that TTX is inaugurated from the microorganisms isolated in the TTX-having organisms. The method used in this research was to drop the egg fluid in media containing KH2PO4 and/or NaCl. For many years, Bacillus species has been studied for their beneficial biological activities. They are good sources of bioactive compounds, notably antibiotics, therapeutic proteins, enzyme inhibitors and pharmacologically active agents.42 Like other Bacillus bacteria, Bacillus amyloliquefaciens is a gram-positive, rod-shaped, catalase-positive, aerobic, and mucoid bacterium. It is able to form endospores, which help it resistant to harsh conditions such as extreme temperature, radiation, pH or harmful chemical agents.42,43 In addition, this bacterium can produce a large number of metabolites and bio-peptide. Furthermore, some studies showed that bacteriocins and lipopeptides which were synthesized non-ribosomally were antimicrobial metabolites produced by Bacillus species which have many desire characteristics including ability to against gram-positive bacteria, Gram-negative bacteria and filamentous fungi.44,45 Moreover, some lipopeptides such as surfactin, iturin and fengycin were isolated and characterized from Bacillus and checked for biocontrol activities against bacteria and fungi.46
This study not only identified Bacillus amyloliquefaciens in TTX production but also firstly announced the existence of this bacterium in Carcinoscorpius rotundicauda. The presence of TTX in bacteria isolated inside Carcinoscorpius rotundicauda contribute to the toxicological figuration in Carcinoscorpius rotundicauda due to the TTX producing ability of endophytic bacteria.47-50 Moreover, the TTX production by bacteria in the peptone containing media might show the similar TTX peaks in HPLC analyses used in Matsumura research,51 therefore, this study used elisa to detect TTX in organisms before purification. After purification, peptone was removed out of TTX extract for elisa, and HPLC analysis to increase the specificity for TTX detection. The study did not detect the toxicity of Carcinoscorpius rotundicauda in mice because the toxicological information was published in our previous study.52 As results of elisa and HPLC analysis, we found that the isolated Bacillus amyloliquefaciens B1 can produce TTX.
The interaction between ligand-protein is involved in many biological functions and consequent pharmaceutical applications.53 Therefore, the binding investigation makes a great effort in recent years, suggesting many theories to develop proposals based on ligand-protein. The study screened the interaction of TTX with the extracellular domain of noncanonic ABC-type transporter from Bacillus amyloliquefaciens that had the crystal structure of YknZ (5F9Q) deposited in PDB. ABC transporter is needed for the substrate translocation across membranes. Some ATPases do not have transport processes but they play a role in RNA translation and DNA repair. Some ABC transporters can uptake or export nutrients, trace metals, biosynthetic precursors, vitamins, drugs, sterols, lipids, metabolites of primary and secondary processes. Other ABC transporters associate drug or chemical resistance. Accordingly, ABC-type transporter from Bacillus amyloliquefaciens was selected to predict the interaction of it with TTX. By molecular docking, ABC-type transporter might interact with TTX via van der Waals, hydrogen bond, and carbon hydrogen bond. Probably, the protein can bind TTX and then the bacterium can resist to TTX and activate gene expression in Bacillus amyloliquefaciens, resulting TTX production in Bacillus amyloliquefaciens B1. Nearby, the study also looked for other membrane proteins of Bacillus amyloliquefaciens, there was no membrane protein of this bacterium that has already existed 3D structures in PDB. However, the study used the other membrane proteins of Bacillus subtilis and Bacillus cereus. These membrane proteins have high similarity to the primary sequence of Bacillus amyloliquefaciens. They might interact with TTX to involve TTX production in Bacillus subtilis and B. cereus which produced TTX in previous studies.54
The study of Bacillus amyloliquefaciens exopolysaccharides (EPS) displayed the abilities in antimicrobial activities to all test pathogens. The inhibition zones were varied by the types of test microorganisms. The negative controls containing distilled water did not show the inhibition zone. In the study, erythromycin was used as positive control because all the bacteria used in the study were inhibited by erythromycin to apply for validating the experimental procedure. Because Staphylococcus aureus and Pseudomonas aeruginosa were resistant to ampicillin when using alone, the study did not use ampicillin although the inhibition may be obtained when ampicillin was combined with other compounds like hypericin.55 For chloramphenicol, this antibiotic can cause toxicity in bone marrow.56 However, the result will give an alteration in therapy when bacteria were not susceptible to ampicillin and chloramphenicol.
EPS in heated and unheated treatments showed insignificant results which meant that EPS is stable in high temperature. EPS from Bacillus amyloliquefaciens was presented to have antimicrobial activities against gram-positive and gram-negative bacteria. EPS of Bacillus amyloliquefaciens had the strongest antimicrobial activities for Staphylococcus aureus but the weakest for Vibrio parahaemolyticus. EPS could inhibit the gram positive bacteria and gram negative bacteria; however, EPS could suppress gram positive bacteria more strongly due to EPS interacted to the bacterial cell walls of and then spoiled the gram positive bacteria much than the gram negative bacteria which do not have cell wall.57 EPS might also bind to receptor and then kill microbes.58 Generally, the interesting mechanisms of EPS in antimicrobial activities are still not clear that should be exploited more. The EPS of Bacillus amyloliquefaciens has not been studied before. The study focused on the antimicrobial activities to clear the reason why there was not many bacterial strains existing in Carcinoscorpius rotundicauda due to the isolated strain could inhibit other bacteria. Moreover, EPS are claimed to be usually biocompatible, edible, and nontoxic to humans and the environment. Therefore, the EPS of Bacillus amyloliquefaciens in this study is suggested to be a source of natural antioxidants with potential value for functional foods or therapeutics. The peak showed EPS contained glucose monomer, pointing EPS was obtained in the study. Surprisingly, EPS produced in Bacillus amyloliquefaciens when cultured in LB medium was lower than MRS medium that was respective to TTX detection in those media. According to the formula of MRS medium,59 peptone, and yeast/meat extracts are the carbon, and nitrogen sources for almost bacterial growth; however, polysorbate 80, a surfactant helps for nutrient uptake in bacteria. Magnesium sulfate and manganese sulfate, dipotassium hydrogen phosphate, triammonium citrate are essential for metabolism. Remarkably, TTX yield increased when EPS production increased in the bacterium in this study. It can be said that EPS has a relation to TTX production in Bacillus amyloliquefaciens that will also be an interesting hypothesis.
In this study, Bacillus amyloliquefaciens B1 isolated from Carcinoscorpius rotundicauda produced TTX. Bacillus amyloliquefaciens B1 produced TTX in MRS medium more highly than LB medium. EPS of Bacillus amyloliquefaciens B1 was detected highly in case of Bacillus amyloliquefaciens B1 cultured in MRS medium. EPS might concern TTX production in Bacillus amyloliquefaciens B1. By docking, TTX could bind to membrane proteins. EPS can kill bacteria bioactivities that will contribute a lot to our life. Therefore, Bacillus amyloliquefaciens B1 isolated from horseshoe crab is suggested to be a new source for pharmaceutical fields.
ACKNOWLEDGMENTS
The authors would like to thank International University (IU), Vietnam National University, Ho Chi Minh City (VNU-HCM) for the laboratory support.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
TN conceptualized the study, provided funding support and proposal investigator. PH supplied crabs and conceptualized the study. BP and YT prepared materials. CL and TN designed experiments. CL and TH performed molecular docking. CL, TD, PL and TN performed experiments. CL, TD and TN performed data analysis. CL and TN wrote the manuscript. All authors read and approved the final manuscript for publication.
FUNDING
None.
DATA AVAILABILITY
The datasets generated and/or analysed during the current study are available from the corresponding author on reasonable request.
ETHICS STATEMENT
Not applicable.
- Zheng R, Guan Q, Zheng M, et al. Yang, Toxin and toxicity identification of mangrove horseshoe crab Carcinoscorpius rotundicauda collected from South China. Toxicon. 2019;161:23-27.
Crossref - Kumar V, Roy S, Sahoo AK, Behera BK, Sharm AP. Horseshoe crab and its medicinal values. Int J Curr Microbiol Appl Sci. 2015;4(2):956-964.
- Walls EA, Berkson J, Smith SA. The Horseshoe Crab, Limulus polyphemus: 200 Million Years of Existence, 100 Years of Study. Rev Fish Sci. 2002;10(1):39-79.
Crossref - Kumar V, Roy S, Sahoo AK, Kumar V. Horseshoe crabs: biomedical importance and its potential use in developing health-care products. Indian J Geo-Mar Sci. 2016;45(10):1234-1244.
- Jang J, Yotsu-Yamashita M. Distribution of tetrodotoxin, saxitoxin, and their analogs among tissues of the puffer fish Fugu pardalis. Toxicon. 2006;48(8):980-987.
Crossref - Tsai YH, Ho PH, Hwang CC, Hwang PA, Cheng CA, Hwang DF. Tetrodotoxin in several species of Xanthid crabs in southern Taiwan. Food Chem. 2006;95(2):205-212.
Crossref - Asakawa M, Matsumoto T, Umezaki K, et al. Toxicity and Toxin Composition of the Greater Blue-Ringed Octopus Hapalochlaena lunulata from Ishigaki Island, Okinawa Prefecture, Japan. Toxins. 2019;11(5):245.
Crossref - Ritson-Williams R, Yotsu-Yamashita M, Paul VJ. Ecological functions of tetrodotoxin in a deadly polyclad flatworm. Proc Natl Acad Sci U S A. 2006;103(9):3176-3179.
Crossref - Yotsu-Yamashita M, Mebs D, Kwet A, Schneider M. Tetrodotoxin and its analogue 6-epitetrodotoxin in newts (Triturus spp; Urodela, Salamandridae) from southern Germany. Toxicon. 207;50(2):306-309.
Crossref - Cheng CA, Hwang DF, Tsai YH, et al. Microflora and tetrodotoxin-producing bacteria in a gastropod, Niotha clathrate. Food Chem Toxicol. 1995;33(11):929-934.
Crossref - Huey RB, Moody WJ. Snake sodium channels resist TTX arrest. Science. 2002;297(5585):1289-1290.
Crossref - Bacchiocchi S, Campacci D, Siracusa M, et al. A Hotspot of TTX Contamination in the Adriatic Sea: Study on the Origin and Causative Factors. Mar Drugs. 2023;21(1):8.
Crossref - Nieto FR, Cobos EJ, Tejada MA, Sanchez-Fernandez C, Gonzalez-Cano R, Cendan CM. Tetrodotoxin (TTX) as a Therapeutic Agent for Pain. Mar Drugs. 2012;10(2):281-305.
Crossref - Gonzalez-Cano R, Ruiz-Cantero MC, Santos-Caballero M, Gomez-Navas C, Tejada MA, Nieto FR. Tetrodotoxin, a Potential Drug for Neuropathic and Cancer Pain Relief? Toxins. 2021;13(7):483.
Crossref - Hagen NA, Cantin L, Constant J, et al. Tetrodotoxin for Moderate to Severe Cancer-Related Pain: A Multicentre, Randomized, Double-Blind, Placebo-Controlled, Parallel-Design Trial. Pain Res Manag. 2017;7212713.
Crossref - Baker-Austin C, Oliver JD, Alam M, et al. Vibrio spp. Infections. Nat Rev Dis Primers. 2018;4:8.
Crossref - Lu Y, Yi R. Bacillus horikoshii, a tetrodotoxin-producing bacterium isolated from the liver of puffer fish. Ann Microbiol. 2009;59:452-458.
Crossref - Yuan Y, Guan F, Yu C, Chen Z, Zhang J. Isolation and Toxigenic Characteristics of a Tetrodotoxin Producing Bacteria. Turkish J Fish Aquat. 2021;22(1):TRJFAS19531.
Crossref - Tu N, Tu Q, Tung H, Hieu D, Romero-Jovel S. Detection of tetrodotoxin-producing Providencia rettgeri T892 in Lagocephalus pufferfish. World J Microbiol Biotechnol. 2014;30(6):1829-1835.
Crossref - Tu N, Huu N, Nghe DV, Kim N. Biological Activities of Tetrodotoxin-Producing Enterococcus faecium AD1 Isolated from Puffer Fishes. Biomed Res Int. 2015;973235.
Crossref - Liu J, Wei F, Lu Y, Ma T, Zhao J, Gong X, Bao B. Production level of tetrodotoxin in Aeromonas is associated with the copy number of a plasmid. Toxicon. 2015;101:27-34.
Crossref - Bacchiocchi S, Campacci D, Siracusa M, et al. Tetrodotoxins (TTXs) and Vibrio alginolyticus in Mussels from Central Adriatic Sea (Italy): Are They Closely Related? Mar Drugs. 2021;19(6):304.
Crossref - Tu N, Vinh D, Anh N. Antimicrobial, Antioxidant and Cytotoxic Activities of Different Fractions Obtained from Endophytic Fusarium solani F01 Isolated in Catharanthus roseus Collected in Vietnam. Indian J Pharm Educ Res. 2022;56(2):461-469.
Crossref - Tu N, Thuy N, Vinh D, Anh N, Ly H. Detection of Antimicrobial, Antioxidant and Cytotoxicity Activities of Fusarium oxysporum F01 Isolated from Catharanthus roseus Collected in Vietnam. J Pure Appl Microbiol. 2021;15(3):1643-1654.
Crossref - Ortega-Morales BO, Santiago-Garcia JL, Chan-Bacab MJ, et al. Characterization of extracellular polymers synthesized by tropical intertidal biofilm bacteria. J Appl Microbiol. 2007;102(1):254-264.
Crossref - Tu N, Dat N, Canh L, Vinh D. Detection of the Potential Inactivation of Tetrodotoxin by Lactic Acid Bacterial Exopolysaccharide. Toxins. 2018;10(7):288.
Crossref - Canh L, Tu Q, Tu N. Factors might increase toxicity of tetrodotoxin. J Chem Pharm Res. 2016;8(8):459-463.
- Huerta-Cepas J, Serra F, Bork P. ETE 3: Reconstruction, analysis, and visualization of phylogenomic data. Mol Biol Evol. 2016;33(6):1635-1638.
Crossref - Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307-321.
Crossref - Cosconati S, Forli S, Perryman AL, Harris R, Goodsell DS, Olson AJ. Virtual screening with AutoDock: theory and practice. Expert Opin Drug Discov. 2010;5(6):597-607.
Crossref - Jaghoori MM, Bleijlevens B, Olabarriaga SD. 1001 Ways to run AutoDock Vina for virtual screenin. J Comput Aided Mol Des. 2016;30:(3)237-249.
Crossref - Morris GM, Huey R, Olson AJ. Using AutoDock for ligand-receptor docking. Curr Protoc Bioinform. 2008;24(1):8.14.1-8.14.40.
Crossref - Xu R, Ma S, Wang Y, Liu L, Li P. Screening, identification and statistic optimization of a novel exopolysaccharide producing Lactobacillus paracasei HCT. African J Microbiol Res. 2010;4(9):783-795.
- Ruhmann B, Schmid J, Sieber V. Methods to identify the unexplored diversity of microbial exopolysaccharides. Front Microbiol. 2015;6:565.
- Dubois M, Gilles KA, Hamilton JK, Rebers PA, Smith F. Colorimetric Method for Determination of Sugars and Related Substances. Anal Chem. 1956;28(3):350-356.
Crossref - Masuko T, Minami A, Iwasaki N, Majima T, Nishimura SI, Lee YC. Carbohydrate analysis by a phenol-sulfuric acid method in microplate format. Anal Biochem. 2005;339(1):69-72.
Crossref - Ismail B, Nampoothiri KM. Molecular characterization of an exopolysaccharide from a probiotic Lactobacillus plantarum MTCC 9510 and its efficacy to improve the texture of starchy food. J Food Sci Technol. 2015;51:4012-4018.
Crossref - Hewetson M, Aaltonen K, Tulamo R, Sankari S. Development and validation of a gas chromatography-flame ionization detection method for quantifying sucrose in equine serum. J Vet Diagn Invest. 2014;26(2):232-239.
Crossref - Mcdermott PF, White DG, Zhao S, Simjee S, Walker RD. Antimicrobial Susceptibility Testing. Preharvest and Postharvest Food Safety: Contemporary Issues and Future Directions. 2008;189.
Crossref - Tu N, Dao N. Detection on Antioxidant and Cytotoxicity Activities of Exopolysaccharides Isolated in Plant-Originated Lactococcus lactis. Biomed Pharmacol J. 2014;7(1).
Crossref - Ghalem BR. Antioxidant and Antimicrobial Activities of Exopolysaccharides from Yoghurt Starter. Adv Biochem Eng. 2017;5(5):97-101.
Crossref - Stein T. Bacillus subtilis antibiotics: structures, syntheses and specific functions. Mol Microbiol. 2005;56(4):845-857.
Crossref - Meena KR, Kanwar SS. Lipopeptides as the antifungal and antibacterial agents: applications in food safety and therapeutics. Biomed Res Int. 2015;473050.
Crossref - Xun-chao C, Hui L, Ya-rong X, Chang-hong L. Study of endophytic Bacillus amyloliquefaciens CC09 and its antifungal cyclic lipopeptides. J Appl Biol Biotechnol. 2013;1(1).
Crossref - Kadaikunnan S, Rejiniemon T, Khaled JM, Alharbi NS, Mothana R. In-vitro antibacterial, antifungal, antioxidant and functional properties of Bacillus amyloliquefaciens. Ann Clin Microbiol Antimicrob. 2015;14:9.
Crossref - Rao BP, Sudharsan K, Sekaran RCH, Mandal AB. Characterization of Exopolysaccharide from Bacillus amyloliquefaciens BPRGS for its Bioflocculant Activity. Int J Sci Eng Res. 2013;4(10):1696-1704.
- Matsumura K. In vivo neutralization of tetrodotoxin by a monoclonal antibody. Toxicon. 1995;33(9):1239-1241.
Crossref - Matsumura K. Reexamination of tetrodotoxin production by bacteria. Appl Environ Microbiol. 1995;61(9):3468-3470.
Crossref - Matsumura K. A Monoclonal antibody against tetrodotoxin that reacts to the active group for the toxicity. Eur J Pharmacol. 1995;293(1):41-45.
Crossref - Matsumura K. Production of tetrodotoxin in pufferfish embryos. Environ Toxicol Pharmacol. 1998;6(4):217-219.
Crossref - Matsumura K. No ability to produce tetrodotoxin in bacteria. Appl Environ Microbiol. 2001;67(5):2393-2394.
Crossref - Canh L, Phuc H, Phong L, Tu N. Conditions involving the toxicity of tetrodotoxin from Carcinoscorpius rotundicauda. World J Pharm Pharm Sci. 2023;12:62-69.
- Salmaso V, Moro S. Bridging Molecular Docking to Molecular Dynamics in Exploring Ligand-Protein Recognition Process: An Overview. Front Pharmacol. 2018;9:923.
Crossref - Magarlamov TY, Melnikova DI, Shokur OA, Gorobets EA. Rapid production of tetrodotoxin-like compounds during sporulation in a marine isolate Bacillus spp. 1839. J Microbiol. 2017;86(2):192-196.
Crossref - Alam ST, Le TAN, Park JS, Kwon HC, Kang K. Antimicrobial Biophotonic Treatment of Ampicillin-Resistant Pseudomonas aeruginosa with Hypericin and Ampicillin Cotreatment Followed by Orange Light. Pharmaceutics. 2019;11(12):641.
Crossref - Wiest DB, Cochran JB, Tecklenburg FW. Chloramphenicol toxicity revisited: a 12-year-old patient with a brain abscess. J Pediatr Pharmacol Ther. 2012;17(2):182-188.
Crossref - Zhou Y, Cui Y, Qu X. Exopolysaccharides of lactic acid bacteria: structure, bioactivity, and associations: a review. Carbohydr Polym. 2019;207:317-332.
Crossref - Abdalla AK, Ayyash MM, Olaimat AN, et al. Exopolysaccharides as Antimicrobial Agents: Mechanism and Spectrum of Activity. Front Microbiol. 2021;12:664395.
Crossref - De Man JC, Rogosa M, Sharpe ME. A Medium for the Cultivation of Lactobacilli. J Appl Bacterial. 1960;23(1):130-135.
Crossref
© The Author(s) 2023. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.