ISSN: 0973-7510
E-ISSN: 2581-690X
The harmless immigration of endophytic microflora in plants and their ability to synthesize various valuable compounds has attracted many researchers to work with plant-microbe interactions and also to exploit them for agricultural and medical applications. This investigation has been carried out to study endophytic bacteria in Biophytum sensitivum (L.) DC by the isolation, characterization and identification based on morphological features, cell characteristics, biochemical tests, plant growth promotion, 16S rDNA sequencing and phylogenetic analysis. Five different bacterial isolates were identified from this study using BLAST analysis of the 16S rDNA sequences and were submitted in GenBank followed by retrieval of accession numbers. The identified bacteria with their accession numbers are Staphylococcus sp. strain (MH050396); Bacillus sp. strain (MH050388); Bacillus cereus strain (MH050384); Bacillus subtilis strain (MH050389) and Bacillus sp. strain (MH050399). All isolates except Bacillus sp. strain (MH050399) produced Indole -3- acetic acid and the highest amount of 14.50µg/ml was obtained from Bacillus subtilis strain (MH050389). All bacterial endophytes reported in this study produced ammonia and siderophore thus indicating their role in plant growth promotion.
Endophytic bacteria, Biochemical Characterization, Plant Growth activity, Phylogenetic analysis.
The plant microbiomes have a very significant contribution in maintaining the growth and existence of their mutualistic partners. This plant-microbe co-evolution and interdependency became more significant area of research with the emergence of Lamarckian concept of the Hologenome theory of Evolution2. Endophytic bacteria are those that can live as asymptomatic colonies inside plant tissues3. The rhizosphere contains diverse variety of bacteria that can gain entry into the plant tissues during favourable conditions. The endophytic bacteria can be obligative or facultative survivals of internal plant tissues4. Bacterial endophytes gain more advantage than the epiphytic bacteria due to their direct contact with host tissues that helps in better communication and survival5. The ecological niche provided by the host plants foster the endophytes in establishing an endosymbiotic relation by providing favourable temperature, osmotic potential and pH. The epiphytic bacteria that are in direct exposure to the changing environmental conditions will not get such benefits. This selects a community of bacterial biome that can colonize endophytically. The symbiont in turn provides the host with various plant growth stimulating compounds like Indole- 3-Acetic Acid (IAA), an auxin that regulates cell division, differentiation and elongation6. They also produce iron chelating compounds like siderophores thus overcoming adverse iron limiting environmental conditions. In return, the endophytes receive shelter and nutrients from the host. Though root zones are the major sites of entry7, endophytic bacteria can also enter through natural wounds or direct entry through aerial openings8 or through stem lenticels9.
Many researchers substantiated the interrelationship between the host and symbiont in different ways. The presence of 14 bacterial endophytes of Curcuma longa L. was categorized based on morphological, biochemical and 16S rRNA sequence analyses10. Seeds and roots of common bean revealed the endophytic colonization of bacteria11. Endophytic bacteria were already reported in the medicinally important plants such as Mentha arvensis, Catharanthus roseus, Stevia rebaudiania and Ocimum sanctum12. Recently, the diversity, colonization capacity and plant growth promoting activity of endophytes have been reviewed in detail13. In this context, Biophytum sensitivum (L.) DC, a highly valuable medicinal herb belonging to the Daspushpam category of Ayurvedically important plant of the family Oxalidaceae, has been selected for the isolation, identification and characterisation of growth promoting endophytic bacteria.
Collection of the plant material and Surface sterilization
Healthy, undamaged and disease-free plants of B. sensitivum, collected from the Botanic Garden of Union Christian College, Aluva, Kerala, India, served as the study material for the isolation and characterisation of endophytic bacteria.
Isolation
Collected plants were rinsed in sterile distilled water followed by its transfer to pre-treatment solution containing 2% dextran for 1min with vigorous shaking without causing damage to the plant. The pre-treated plant was carefully washed with sterilized double distilled water (DDW) 4-5 times, after which the plant was washed in 0.1% mercuric chloride (HgCl2) for 1min followed by again washing in DDW five times, 2min for each wash. It was then rinsed in 70% ethanol for 1min followed by three rounds of wash in DDW for 2min each. From the final wash, 0.1ml was poured into a 2.8% nutrient agar plate which was used as control. The surface sterilized plant material was crushed with phosphate buffer saline and was diluted serially up to 10-3 from which 0.1ml was plated on to nutrient agar plates and incubated at 28±2°C for 48hrs14. Control was observed first. Absence of bacterial growth on the control plates and efficient growth on the nutrient plates indicated complete surface sterilization of the material. Morphologically different bacterial colonies were sub cultured in fresh nutrient media followed by storage in nutrient agar for further analysis.
Preliminary characterisation of endophytic bacteria
The bacterial colonies obtained were subjected to phenotypic studies by morphological and biochemical characterisation. Cultural properties like shape, elevation, edge, colour and texture of bacterial colony and microscopic examination involved motility testing, gram staining and shape of cells. Based on morphological features, the isolated bacteria were classified into five groups such as BS1, BS2, BS3,BS4 and BS5. Further experiments were conducted in these groups only. Biochemical characterisation was performed by citrate utilization, hydrogen sulphide, indole, litmus milk decolourization, nitrate reduction, oxidation-fermentation(O-F), phenylalanine deaminase, urease, Voges- Proskauer(V-P) and methyl red tests.
Screening for Plant Growth Activity
IAA production
The bacterial isolates were inoculated into 10mL of nutrient broth supplemented with 0.2% (v/v) of L- tryptophan and incubated for 10 days at 28°C. This culture was then centrifuged at 3000rpm for 20min and the supernatant collected15. 1mL of the supernatant was mixed with 2mL of freshly prepared Salkowski reagent (12 g of FeCl3/L, 7.9 M of H2SO4) and incubated in dark for 30min. The development of red colour would indicate a positive result.
Quantitative Estimation of IAA
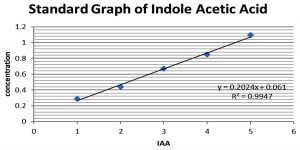
For colourimetric estimation the bacterial isolates were inoculated into 200mL of nutrient broth supplemented with 0.2% v/v of L-tryptophan and incubated for 10 days at 28°C. After incubation, the cell free extract was collected by centrifugation at 3000rpm for 20 min. The supernatant was acidified to pH 2.5 with 1N HCl and was extracted twice with ethyl acetate. The extracted ethyl acetate fraction was vacuum dried at 40°C. The residue was resuspended in 3ml ethyl acetate, from which 1ml of the supernatant was mixed with 2ml of freshly prepared Salkowski reagent and incubated in dark for 20min. The absorbance was read at 530nm. The concentration of IAA present in each sample was calculated using standard curve plotted against varying concentrations of IAA.(Fig. 1)
Ammonia production
To 10 ml bacterial suspension in peptone water, Nessler’s reagent was added and the development of brown to yellow colour would indicate the production of ammonia.
Phosphate solubilization
Phosphate solubilisation was tested using standard protocols14. Pikovskaya medium containing 2.4mg/mL bromophenol was used for the screening and the isolates were incubated for 48hrs. Positive results would indicate a yellow zone around the colony due to the utilization of tricalcium phosphate in the medium.
Siderophore production
1ml of 2% aqueous Fe Cl3 was added to 1ml of the culture supernatant obtained from King’s medium, Development of orange or red-brown colouration would indicate the presence of siderophore.
ACC deaminase production
The isolates were inoculated to DF salt minimal medium amended with 0.2% ammonium sulphate (w/v). The bacterial growth in this media after 2 days of incubation was considered as positive result.
Identification of endophytic bacteria by 16S rRNA sequencing
Genomic DNA of the five selected strains were isolated by NucleoSpin® Tissue kit following the manufactures protocol. The DNA quality and purity were checked using agarose gel electrophoresis in 0.8% gel. The gels were visualized and analysed using Gel documentation system (Bio-Rad). PCR amplification was carried out in a 20µl reaction volume with 1X PCR buffer (100mM Tris HCl , pH-8.3; 500mM KCl), 0.2mM each dNTPs (dATP, dGTP, dCTP and dTTP), 2.5mM MgCl2, 1 U AmpliTaq Gold DNA polymerase enzyme, 0.1 mg/ml BSA, 4% DMSO, 5pM of forward and reverse primers and template DNA. The primers used for amplification of part of 16S rRNA were 16SF(5’CAGGCCTAACACATGCAAGTC3’) and 16SR (5’GGGCGGWGTGTACAAGGC). The PCR amplification was carried out for 35 cycles in a thermal with initial denaturation at 95°C for 5.00 min followed by 35 cycles at 95°C for 30 s, annealing at 60°C for 40s and extension at 72°C for 60s and one cycle of final extension for 7min at 72°C. Sequencing reaction was done using the BigDye Terminator v3.1 Cycle sequencing Kit (Applied Biosystems, USA) following manufactures protocol. The sequence quality was checked using Sequence Scanner Software v1 (Applied Biosystems) and the quality check was done using Geneious Version R6. The DNA sequences were compared with the Genbank database using BLASTN software16. The nucleotide sequences of the partial 16S rDNA gene segments were deposited in GenBank and their accession numbers were obtained. With the above sequences a phylogenetic tree was drawn using MEGA X17 (Version 10.1.7) and the evolutionary history was inferred using Neighbour-Joining method with 1000 bootstrap replicates18.
Endophytic bacteria capable of producing various compounds with therapeutic applications can be considered as a potential alternative to the natural products. With exponential growth rate of population and declining food availability, plant growth promoting endophytic bacteria became potent area of research. Endophytic bacteria can be used as a possible eco-friendly option for enhancing productivity and growth of plants. Experimentation using HgCl2 and ethanol as surface sterilant displayed satisfactory results which was evident while observing the control that showed no bacterial colonies. Five distinct bacterial colonies were identified by morphological characteristics, biochemical tests and 16S rDNA analysis. Based on this the bacteria were named from BS1, BS2, BS3, BS4 and BS5. Visual morphological studies on the cultural properties of the isolates revealed that all isolates were irregular in shape showing raised elevation except BS4 with umbonate elevation. All the isolates had undulate edge except BS3 which showed entire edge. A smooth texture was observed for all the isolates with pale white to yellow colouration. All the bacteria were gram positive and except BS1 all others were motile and rod shaped. The biochemical and physiological characterization of the isolates were summarised in Table 1. From this it was evident that all isolates produced nitrate and indole and fermented dextrose. BS3, BS4 and BS5 utilised citrate as the main carbon source while BS1 and BS2 were citrate negative. Except BS1, all other isolates decolourised litmus milk and fermented methyl red, thus indicating their ability to transform the components of milk and production of stable acids during fermentation respectively. On the contrary, all isolates showed negative results to urease production, VP test, phenylalanine deaminase reduction test, CO2 production and H2S production.
Table (1):
Screening for Biochemical Characteristics of the endophytic isolates.
| Isolates | Citrate Utilization | Klingler iron agar test | Indole Production | Litmus milk decolourization | Nitrate reducing | O- F | PAD | Urease | VP | MR | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C | S | Fermentator | ||||||||||
| BS1 | – | – | – | DF | + | – | + | F | – | – | – | – |
| BS2 | – | – | – | DF | + | + | + | NF | – | – | – | + |
| BS3 | + | – | – | DF | + | + | + | F | – | – | – | + |
| BS4 | + | – | – | DF | + | + | + | F | – | – | – | + |
| BS5 | + | – | – | DF | + | + | + | F | – | – | – | + |
Symbols and abbreviations: DF-Dextrose Fermentators; F-Fermentators; NF-Non fermentators
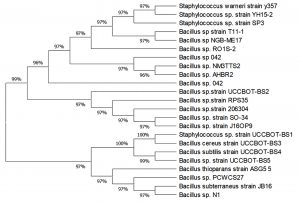
Screening for plant growth promotion activities of the isolates as exhibited in Table 2 revealed that BS4 produced highest amount of IAA (14.50µg/ml) whereas BS1, BS2 and BS3 produced 12.6 , 11.93, and 11.13µg/ml IAA respectively. But BS5 produced the least amount of IAA (8.56µg/ml). However, all the isolates produced ammonia and siderophore. 16S rDNA sequencing was done to identify the bacterial isolates. The sequences were compared with Genbank database and species identification was done based on the results of the BLAST analysis. Accordingly, the five different isolates were named as Staphylococcus sp. strain (BS1), Bacillus sp. strain (BS2), Bacillus cereus strain (BS3), Bacillus subtilis strain (BS4) and Bacillus sp. strain (BS5). The sequences were submitted in the Genbank and corresponding accession numbers were obtained such as MH050396 (BS1) with 1,122bp long sequence, MH050388(BS2) with 1,118bp long sequence, MH050384(BS3) with 1,121bp long sequences, MH050389(BS4) with 1,114bp long sequence and MH050399(BS5) with 1,125bp long sequence as given in Table 3. A phylogenetic tree was constructed using MEGA X that indicated their closeness to other taxonomically similar microorganisms Fig. 2. The phylogenetic tree revealed distinct clustering of BS1, BS3, BS4 and BS5 which is an indication of their close relatedness whereas BS2 formed a separate clade. The optimal tree was with the sum of branch length = 63.21930360. The evolutionary distances were computed using Maximum Composite Likelihood method19 and were in the units of the number of base substitutions per site. The proportion of sites where at least one unambiguous base is present in at least one sequence for each descendent clade is shown next to each internal node in the tree. This analysis involved 23 nucleotide sequences. All ambiguous positions were removed for each sequence pair (pairwise deletion option). There were a total of 1125 positions in the final dataset.
Table (2):
Screening for Plant growth promotion activity.
Isolates |
IAA Production |
Ammonia production |
Siderophore production |
Phosphate solubilization |
ACC deaminase activity |
|---|---|---|---|---|---|
BS1 |
+ |
+ |
+ |
– |
– |
BS2 |
+ |
+ |
+ |
– |
– |
BS3 |
+ |
+ |
+ |
– |
– |
BS4 |
+ |
+ |
+ |
– |
– |
BS5 |
– |
+ |
+ |
– |
– |
Table (3):
BLAST analysis and sequence submission.
Source Organism |
Bacterial strain |
Accession number |
Nearest phylogenetic neighbour |
Homology |
|---|---|---|---|---|
Biophytum sensitivum |
BS1 |
MH050396 |
Staphylococcus sp. strain Y117(JX077097.1) |
97% |
Biophytum sensitivum |
BS2 |
MH050388 |
Bacillus sp. strain YNA54 (JN867119.1) |
99% |
Biophytum sensitivum |
BS3 |
MH050384 |
Bacillus cereus strain EP218 (MG778892.1) |
98% |
Biophytum sensitivum |
BS4 |
MH050389 |
Bacillus subtilis strain EAF42 (MG593998.1) |
98% |
Biophytum sensitivum |
BS5 |
MH050399 |
Bacillus sp. strain KUDC1744(KC414723.1) |
98% |
The study on isolation and characterisation of bacterial endopytes in B. sensitivum has been undertaken to prove the ability of such bacteria in regulating plant growth and development. The most important prerequisite for endophyte isolation is the standardisation of an effective surface sterilization of the plant material. In the present experiment successful surface sterilization has been achieved using HgCl2 and ethanol and similar results were reported earlier12. Even though surface sterilization was achieved, successful growth of endophytes depends on whether the chemical agent adversely affected the endophytic microbiome of that plant or not20,21. From B. sensitivum bacterial endophytes such as Staphylococcus sp. strain, Bacillus sp. strain, Bacillus cereus strain, Bacillus subtilis strain and Bacillus sp. strain were isolated and identified based on morphological, biochemical and molecular methods. It is a routine practice in microbiology to identify bacteria based on visual morphology and biochemical tests. Even after these regular experimentations, the identity of bacteria may become a problem that cannot be solved by Bergey’s manual of bacteriology. Hence molecular characterisation using 16S rDNA sequencing was employed for the identification of endophytes from B. sensitivum. 16S rDNA was highly conserved, distributed unvaryingly in the genome of microbes and less influenced by horizontal gene transfer22. However certain variable regions in 16S rDNA are prone to variations. This variability can be used as a tool for the classification of endophytes by designing PCR primers for conserved regions23. Based on 16S rDNA sequencing of endophytic bacteria in B. sensitivum we obtained five-gram positive bacteria that were phylogenetically classified using BLAST analysis and MEGA X software. Among the different endophytes Bacillus subtilis strain and Bacillus cereus strain were previously reported10,24,25. Based on molecular analysis, a phylogenetic tree was constructed revealing the evolutionary relationship between the isolates and other taxonomically related microorganisms. Two major clades were formed with an inferior clade that separated the Staphylococcus sp. strain from all the four isolates, clustered away from BS1. Similar to the present study, various evolutionary studies on endophytic bacteria residing in several plants have been performed by using phylogenetic tree26. Costa et al.27 isolated bacterial endophytes of the genera Staphylococcus and Bacillus from the leaves of Phaseolus vulgaris. The presence of Bacillus sp. has been previously reported from the roots of Cassia tora L28. Endophytic bacteria were previously reported as protagonist in regulating plant growth, development as well as survival. Etesami et al.29 was of opinion that the most substantial plant growth promoting factor was IAA, contributing to the successful colonization of bacteria inside the plant tissues. In B. sensitivum, all the endophytic strains except Bacillus sp. strain (BS5) produced substantial amount of IAA when tryptophan was present. BS4 produced the highest amount of IAA (14.50µg/ml). This could be a major contributing factor towards increased plant growth promotion30. Similar results were reported with Bacillus cereus (ECL1) and Bacillus sp. strain (ECL3) as producers of IAA in the rhizome of Curcuma longa L10. Kumar et al.28 also reported the production of IAA by bacterial endophytes such as Bacillus subtilis sp. strain that promotes plant growth. In the present study, all the isolated strains produced ammonia which can be considered as a contributing trait for plant growth promotion31. A study on the characterisation of the bacterial endophytes from both leguminous and non-leguminous plants revealed a total of 44 bacterial endophytes that exhibited the ability to produce ammonia32. Research reports documented by Ngoma et al.33 revealed that except Stenotrophomonas maltophilia, all the seven bacterial endophytes produced ammonia which indicated their significance in plant development. Screening for siderophore production showed a positive result by all the isolates which may be advantageous to iron deficient plants by chelating Fe3+ 34. Similar results were shown during the study conducted by Brigido et al.35 in which 33.3% of the test bacterial endophytes isolated from Chickpea showed the ability to synthesize siderophores, thus playing an indirect role in inhibiting the multiplication of phytopathogens by sequestrating iron. The present investigation revealed that none of the isolates were able to synthesize phosphate and ACC deaminase. On the contrary, Oteino et al.36 verified a moderate to high profile of phosphate solubilization capacities in which Pseudomonas strains L111, L228 and L321 showed good phosphate solubilizing ability and the L321strain expressing the trait in phosphate limiting conditions. Contrasting results were also shown by various researchers for the production of ACC deaminase substantiating their ability to promote plant growth37.
The results from the study demonstrated the successful isolation of five endophytic bacteria associated with B. sensitivum. These isolates confer plant growth promotion by producing many compounds like IAA, siderophore and ammonia. Among the isolates BS4 was found to produce the highest amount of IAA which was quantitatively confirmed. 16S rDNA sequencing was conducted and the corresponding sequences of all the five isolates were submitted in GeneBank Database and obtained unique accession numbers. This study demonstrated that plant growth promoting trait of the endophytic bacteria could be exploited in future as a contributing factor for the growth of the economically important plants. Thus, future experimentation can be directed towards the goal to exploit plant growth promoting traits of the endophytic isolates to increase the production of good quality crops. This bioresource can be considered as a safer, eco-friendly and sustainable alternative in amending the agricultural crop yields when compared to chemical fertilizers that often harm the environment and humans.
ACKNOWLEDGMENTS
The authors are grateful to the Department of Botany Union Christian College, Aluva and Rajiv Gandhi Centre for Biotechnology, Trivandrum for the facilities provided.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed above have made a direct involvement in the work and approved it for publication.
FUNDING
None.
ETHICS STATEMENT
This article does not contain any study involving human participants or animals.
AVAILABILITY OF DATA
All the data obtained in the study are incorporated in the manuscript.
- Berendsen RL, Pietese CMJ, Bakker P. The rhizosphere microbiome and plant health. Trends in Plant Sci.,2012; 17: 478-486.
Crossref - Rosenberg E, Sharm G, Zilber-Rosenberg I. The hologenome theory of evolution contains Lamarckian aspects within a Darwian framework. Environ Microbiol.,2009, 11(12): 2952-62.
Crossref - Arora J, Ramawat KG. An Introduction to Endophytes. Sustainable Develop and Biod. 2017; 9: 1-23.
Crossref - Kado CI. Plant Pathogenic bacteria, pp 659-674. In: Balows A, Truper H G, Dworkin M, Harder W, Schleifer KH (eds). The prokaryotes Vol I, New York, 1992.
- Hallmann J, Quadt-Hallmann A, Mahaffee WF, Kloepper JW. Bacterial endophytes in agricultural crops. Can J Microbiol., 1997a; 43(10): 895-914.
Crossref - Taghavi S, Garafola C, Monchy S, Newman L, Hoffman A, Weyens N, Barac T, Vagronsveld J, Van der Lelie D. Genomic survey and characterization of endophytic bacteria exhibiting a beneficial effect on the growth and development of popular trees. Appl Environ Microbiol., 2009; 75(3): 748–757.
Crossref - Quadt- Hallmann A, Kloepper JW. Immunological detection and localization of the Cotton endophyte Enterobacter asburiae JM22 in different plant species. Can. J. Microbial., 1996; 42: 1144 -1154.
Crossref - Sharrock KR, Parkes SL, Jack HK, Rees-George J, Hawthorne BT. Involvement of bacterial endophytes in storage rots of buttercup squash (Cucurbita maxima. D.hybrid Delicia). New Zealand J. Crop Horticultural Sci., 1991; 19: 157–165.
Crossref - Kluepfel DA. The behavior and tracking of bacteria in the rhizosphere. Ann. Rev. Phytopath., 1993; 31: 441–472.
Crossref - Kumar A, Singh R, Yadav A, Giri DD, Singh KP, Pandey DK. Isolation and bacterial characterization of bacterial endophytes of Curcuma longa. L. Biotech., 2016; 6: 60.
Crossref - Lopez-Lopez A, Rogel MA, Ormeno-orillo Martinez-Romero J. Phaseolus vulgaris seed born endophytic community with novel bacterial species such as Rhizobium endophytum sp. Applied Microb., 2010; 33: 322-327.
Crossref - Anjum N, Chandra R. Endophytic bacteria: Optimization of isolation procedure from various medicinal plants and their preliminary characterization. AJPCR., 2015; 8: 233-238.
- Maela PM, Serepa – Dlamini MH.Current Understanding of Bacterial Endophytes, Their Diversity, Colonization and Their Roles in Promoting Plant Growth. Appli Microbiol., 2019; 5: 1.
- Jasim B, Jimtha JC, Mathew J, Radhakrishnan EK. Isolation and characterization of plant growth promoting endophytic bacteria from rhizome of Zingiber officinale. Biotech., 2013; 4: 197-204.
Crossref - Rahman A, Sitepu R, Tang S Y, Hashidoko Y. Salkowski’s reagent test as a primary screening index for functionalities of Rhizobacteria isolated from wild dipterocarp saplings growing naturally on medium-strongly acidic tropical peat soil. Biosci Biotech Bioch., 2010; 74: 2202-2208.
Crossref - Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 1997; 1: 3389-3402.
Crossref - Kumar S, Stecher G, Li M, Knyaz C and Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol., 2018; 35: 1547-1549.
Crossref - Saitou N and Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol., 1987; 4: 406-425.
- Tamura K, Nei M and Kumar S. Prospects for inferring very large phylogenies by using the neighbour-joining method. PNAS., 2004; 101: 11030-11035.
Crossref - Mercado-Blanco J, Lugtenberg BJJ Biotechological Applications of Bacterial Endophytes. Current Biotech.,2014; 3: 60-75.
Crossref - Eevers N, Gielen M, Sanchez-Lopez A, Jaspers S, White JC, Vangronsveld J, Weyens N. Optimization of isolation and cultivation of bacterial endophytes through addition of plant extract to nutrient media. Microbial Biotech., 2015; 8: 707–715.
Crossref - Daubin V, Moran NA, Ochman H. Phylogenetics and the cohesion of bacterial genomes. Science, 2003; 301: 829-832.
Crossref - Head IM, Saunders JR, Pickup RW. Microbial evolution, diversity and ecology: A decade of ribosomal RNA analysis of uncultivated microorganisms. Microb Ecol., 1998; 35: 1-21.
Crossref - Deng Y, Chen H, Li C, Xu J, Qi Q, Xu Y, Zhu Y, Zheng J, Peng D, Ruan L, Sun M.Endophyte Bacillus subtilis evade plant defence by producing lantibiotic subtilomycin to mask self-produced flagellin. Commun Bio., 2019; 2.
Crossref - Urumbil SK, Anilkumar M. Isolation, Identification and characterisation of plant growth promoting bacterial endophytes from Emilia sonchifolia (Linn.) DC. Asian Jr. of Microbiol. Biotech. Env. Sc.,2019; 21: 648-655.
- Etminani F, Harighi B.Isolation and Identification of Endophytic Bacteria with Plant Growth Promoting Activity and Biocontrol Potential from Wild Pistachio Trees. Plant Pathol J., 2018; 34: 208-217.
Crossref - Costa LEO, Vieira de Queiroz M, Borges AC, Alencar de Moraes C, Fernandes de Araujo E. Isolation and Characterization of endophytic bacteria isolated from the leaves of the common bean. Braz J Microbiol., 2012; 1562-1575.
Crossref - Kumar V, Kumar A, Pandey KD and Roy BK. Isolation and characterization of Bacterial endophytes from the roots of Cassia tora. L. Ann. Microbiol., 2015; 65: 1391-1399.
Crossref - Etesami H, Alikhani HA, Hosseini HM. Indole-3-acetic acid (IAA) production trait, a useful screening to select endophytic and rhizosphere competent bacteria for rice growth promoting agents. Methods X., 2015; 2: 72–78.
Crossref - Shi Y, Lou K, Li C. Isolation, quantity distribution and characterization of endophytic microorganisms within sugar beet. Afr. J. Biotechnol., 2009; 8: 835-840.
Crossref - Wani P, Khan M, Zaidi A. Co-inoculation of nitrogen-fixing and phosphate-solubilizing bacteria to promote growth, yield and nutrient uptake in chickpea. Acta Agronom Hungarica., 2007; 55: 3.
Crossref - Zaghloul R, Abou-Aly H, Tewfike TA, Ashry N. Isolation and Characterization of Endophytic Bacteria Isolated from Legumes and Non-Legumes Plants in Egypt. Journal of Pure and App Microb., 2016; 10:277-290.
- Ngoma L, Esau B, Babalola BB Isolation and characterization of beneficial indigenous endophytic bacteria for plant growth promoting activity in Molelwane Farm, Mafikeng, South Africa. Afr. J. Biotechnol., 2013; 12: 4105-4114.
- Khan Z, Doty SL. Characterization of bacterial endophytes of sweet potato plants. Plant Soil., 2009; 322: 197-207.
Crossref - Brigido C, Singh S, Menendez E, Tavares MJ, Glick BR, Felix MR, Oliveira S, Carvalho M. Diversity and Functionality of Culturable Endophytic Bacterial Communities in Chickpea Plants. Plants.,2019; 8(2):42
Crossref - Oteino N, Lally RD, Kiwanuka S, Lloyd A, Ryan D, Germaine KJ, Dowling DN. Plant growth promotion induced by phosphate solubilizing endophytic Pseudomonas isolates. Front. Microbiol., 2015; 6: 745.
Crossref - Khan AL, Halo BA, Elyassi A, Ali S, Al-Hosni K, Hussain J, Al-Harrasi A, Lee I-J.Indole acetic acid and ACC deaminase from endophytic bacteria improves the growth of Solanium lycopersicum. Electron. J. Biotechnol., 2016; 21: 58–64.
Crossref
© The Author(s) 2020. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.