Mausam1, P K Ray1, A Dey1, S mohanty1, P Kaushik2,Anjay2, Manju Sinha3 and S K Gupta1*
2 Department of Veterinary Public Health, Bihar Veterinary College, Patna – 800 014, Bihar, India, 3 Institute of Animal Health and Production, Patna – 800 014, Bihar, India.
ABSTRACT
The aim of this study was to know the prevalence of Staphylococcus aureus in bovine milk samples and to evaluate the occurrence of methicillin-resistant Staphylococcus aureus (MRSA) among S. aureus isolates. A total of 150 bovine milk samples were analysed which showed the prevalence of S. aureus in 56.67% of samples. The mecA gene detection by PCR among S. aureus revealed the distribution of MRSA in 29.33% of bovine milk samples. Antibiotic sensitivity pattern reveals that 86.36 and 95.45 percent MRSA isolates were resistant to penicillin and ampicillin respectively, whereas 80.49 and 87.80 percent non-MRSA isolates were found to be resistant to penicillin and ampicillin antibiotics respectively.
Keywords
Staphylococcus aureus, bovine milk, MRSA, mecA and ABST.
INTRODUCTION
Staphylococcus aureus is recognized as a pathogen of animals and humans responsible for causing a wide variety of diseases1. It colonizes the skin and mucosae causing suppurative disease, arthritis, urinary tract infections and clinical or subclinical mastitis in ruminants2, 3. Presence of S. aureus on the skin and mucosae of food animals and their frequent association with mastitis, often leads to contamination of milk4 which may result in food poisoning in human beings. Preformed enterotoxins (SEs) of S. aureus are responsible for causing food poisoning and this ranks third among reported food-borne illnesses in the world5.
Recently, there has been increased concern about emergence of antibiotic resistant strains of S. aureus. Development of resistance has been attributed to the extensive therapeutic use of antimicrobials or to their administration as growth promoters in food animal production6. Methicillin-resistant Staphylococcus aureus (MRSA) are the strains of S. aureus that are resistant to all available penicillins and other â-lactam antimicrobial drugs and it was first reported in 19617, 8. . Resistance to methicillin is due to the acquisition of the mecA gene, which encodes PBP2a, an alternative protein necessary for building of the bacterial cell wall. PBP2a is encoded by the mecA gene located in the mobile genetic element called Staphylococcal cassette chromosome (SCCmec)9.
The strains of MRSA that affect animals were referred as LA-MRSA (Livestock-associated MRSA)9. More recently, LA-MRSA has emerged as an organism of zoonotic importance and has been isolated from livestock such as pigs, cattle, poultry etc10. However, the first evidence of direct MRSA transmission between cattle and humans was reported by Juhasz-Kaszanyitzky et al.11. There is now increasing concern about the public health impact of MRSA associated with food producing animals12. MRSA strains have been isolated in many countries from cows’ or small ruminants’ milk and various dairy products13. Thus this study was aimed to determine the prevalence of S. aureus and MRSA in bovine milk from different districts of Bihar, India and to generate antibiotic sensitivity profile of isolates.
Materials and Methods
Sample collection
A total of 150 fresh bovine milk samples were collected from different districts of Bihar during January to March, 2016 by the standard sampling methods. The samples were collected from Patna (128), Khagaria (12) and Madhepura (13) district of Bihar and transported to the laboratory under cold conditions.
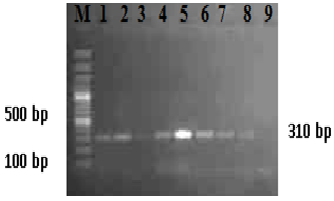
Fig. 1. PCR amplification of mecA gene of methicillin resistant S. aureus
M: 100 bp plus DNA ladder
L1-L8: Positive amplicons of mecA gene of sample isolates
L9: Negative control
Bacterial isolation and biochemical characterization
The milk samples were processed for enrichment by inoculation of 0.5 ml in 10 ml trypticase soy broth with 10% NaCl followed by overnight incubation at 37 °C. Selective plating was performed using Mannitol salt agar (MSA) (HiMedia, India) with overnight incubation at 37°C. Typical yellow colour (mannitol fermentor) colonies on MSA were examined by Gram staining and biochemical tests.
Genomic DNA isolation
The genomic DNA of Staphylococcus aureus was isolated by using the method described by Wilson14 with some modifications. Ten ml of overnight grown bacterial culture was centrifuged at 8000 rpm for 15 minutes and supernatant was discarded. The pellet was washed with normal saline and centrifuged at 8000 rpm for 15 minutes. 0.5 ml of 10mM Tris HCl (pH 8) and lysozyme (2.5 mg/ml) was added and incubated at 37°C for 1 to 2 hours. 6 µl Proteinase K (10 mg/ml) and 30 µl SDS (20%) were added and mixed thoroughly. The mixture was then incubated at 37°C for 60 mins. 100 µl 5 M NaCI and 80 µl CTAB/NaCI solution were added and incubated at 65°C for 10 minutes. Chloroform/isoamyl alcohol (24:1) and phenol/chloroform/isoamyl alcohol (25:24:1) extraction was performed consecutively and DNA was precipitated using 0.7 volumes isopropanol. Finally, the DNA pellet was washed twice with 70% ethanol and dissolved in TE buffer. The DNA was stored at -20oC for future use.
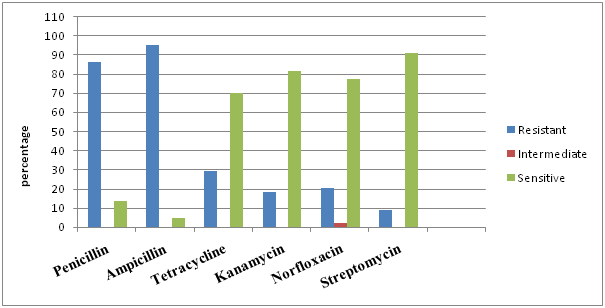
Fig.2. Antibioticsensitivty profile of methicillin resistant S.anreus lsolates from bovine milk
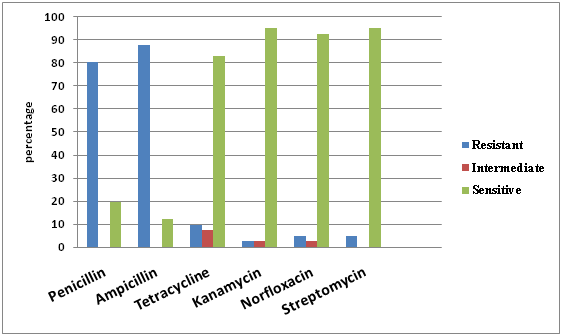
Fig. 3. Antibiotic sensitivity profile of non-MRSA isolates from bovine milk
PCR cyclic conditions for amplification of mecA gene
A polymerase chain reaction (PCR) assay targeting methicillin resistant ‘mecA’ gene was used for detection of methicillin resistant S. aureus as per the method described by Braoios et al.15 with slight modifications. The PCR was performed in a total reaction volume of 25 µl containing 2.5 µl of 10X DreamTaq buffer, 1 µl of 10 mM of each dNTP, 10 pmol of each primer (F: GTAGAAATGACTGAACGTCCGATAA and R: CCAATTCCACATTGTTTCGGTCTAA)15, 1 U Taq polymerase (Thermoscientific), 2 µl of bacterial DNA and nuclease-free water up to 25 µl. The PCR amplification was performed in a thermal cycler (Peqlab, Germany) with initial denaturation at 95°C for 5 min, followed by 35 cycles of denaturation at 95°C for 30 s, annealing at 50°C for 30 s and extension at 72°C for 45s. Final extension was carried out at 72°C for 10 min. The products were electrophoresed in 1% agarose gel (Amresco) stained with ethidium bromide (0.5 µg/ml) and image was documented in a gel documentation system (BioRad).
Antibiotic sensitivity profiling of isolates
The antibiotic sensitivity test of isolates was performed according to protocol described by Clinical Laboratory Standards Institute16. The antibiotic discs were impregnated with penicillin (10µg), ampicillin (10µg), tetracycline (30µg), kanamycin (30µg), norfloxacin (10µg) and streptomycin (10µg) (HiMedia, India). About 100 µl of the inoculums (bacterial culture grown in nutrient broth) was spread over Mueller Hinton agar plate with L- spreader and antibiotic discs were placed onto the plate using sterile forceps. The plates were incubated at 37°C for 24 hrs and observed for zone of inhibition. The results were categorized as resistant, intermediate and sensitive as per CLSI guidelines.
Results and Discussion
Isolation and biochemical identification of S. aureus on MSA from bovine milk samples revealed a total prevalence of S. aureus as 56.67% (85/150) comprising of 54.69% (70/128), 75% (9/12) and 46.15% (6/13) from Patna, Khagaria and Madhepura district of Bihar, India respectively. The findings show a high rate of prevalence in bovine milk since the samples were collected randomly from different animals.
The higher prevalence of S. aureus (~57%) in bovine milk, indicating the risk for development of mastitis in the animals harbouring the organisms or may also attribute to the lack of hygiene and sanitation in the farm or their handlers. The present finding is in concordance with the findings of Sudhan et al.17, who reported a prevalence of 56% S. aureus in India. Similar finding with slightly lower rate of prevalence (50-53%) in milk samples were also reported from different parts of the world18, 19. In contrast to the present finding, a lower prevalence of 28-34% in bovine milk sample was also reported by Abera et al.20 and Joshi et al.21.
The PCR targeting mecA gene was also standardized for rapid detection and confirmation of MRSA. The assay yielded a specific amplicon of a 310 bp (fig. 1). All the isolates confirmed biochemically were tested for amplification of mecA gene by PCR. This revealed the prevalence of MRSA in 29.33% (44/150) of the bovine milk samples. The distribution of mecA gene in the samples belonged to different district of Bihar comprising of 28.12% (36/128), 38.46% (5/13) and 23.08% (3/12) from Patna, Khagaria and Madhepura district, respectively.
This study showed a higher prevalence of MRSA (~29%) in bovine milk that was found in accordance with the findings of Mirzaei et al.22 and Alian et al.23 who reported a prevalence of 29% and 28.3% MRSA in raw milk, respectively. In contrast to the finding of present study a lower prevalence of 0-25% was also reported by others24, 25, 26. The higher prevalence of MRSA in the present study may be due to indiscriminate use of beta-lactam antibiotics as the drug of choice for the treatment of subclinical and clinical mastitis in animals.
The antibiotic sensitivity test was performed against penicillin, ampicillin, tetracycline, kanamycin, norfloxacin and streptomycin. All the isolates (85) were grouped separately into mecA negative (non-MRSA) (41) and mecA positive (MRSA) (44) isolates and result were interpreted accordingly. The antibiotic sensitivity study of S. aureus showed that 80.49 and 87.80 percent non-MRSA isolates were found to be resistant to penicillin and ampicillin antibiotics respectively, while, 2-9% with tetracycline, kanamycin, norfloxacin and streptomycin (fig. 2). In comparison to this MRSA isolates showed a resistance of 86.36 and 95.45 percent to penicillin and ampicillin respectively while, 20-29% to tetracycline and norfloxacin and 9-18% to streptomycin and kanamycin (fig. 3).
The present study showed high resistance of S. aureus to penicillin and ampicillin. This is in accordance with the findings of Abebe et al.27 and Abera et al.28 who reported resistance of S. aureus to penicillin 94% and 94.4% respectively. The results were in accordance with findings of earlier studies from different countries29, 30 suggesting a possible development of resistance from prolonged and indiscriminate usage of antimicrobials. Resistance to penicillin is a great concern since this antibiotic represents the main antibiotic groups recommended for Staphylococcal mastitic infection. The regular use of antibiotics for the treatment of animals might result in the spread of resistant strains as resistance is mediated by plasmids and transposons which can pass from one organism to others.
Conclusions
S.aureus and MRSA is widely prevalent in bovine milk as shown in the present study. With increase in resistance for common antibiotic used in the field condition, the potential of alternative antibiotics found effective in in vitro antibiotic sensitivity test may be considered for treatment against mastitis. The emergence of MRSA in bovine can also pose a potential risk of its transmission from animal to human.
Acknowledgement
The Authors thank Director, ICAR-Research Complex for Eastern Region for providing necessary facilities and funds to carry out the present work.
References
- Bardiau, M., Yamazaki1, K., Duprez, J.N., Taminiau, B., Mainil, J.G. and Ote, I. Genotypic and phenotypic characterization of methicillin resistant Staphylococcus aureus (MRSA) isolated from milk of bovine mastitis. Lett. Appl. Microbiol., 2013; ISSN 0266-8254.
- Asperger, H., Zangerl, P. Staphylococcus aureus. In: Encyclopaedia of dairy sciences. Roginski, H., Fuquay, J.W., Fox, P.F. eds. Academic Press, San Diego, pp. 2563–2569.
- Petridis, I.G., Mavrogianni, V.S., Gougoulis, D.A., Amiridis, G.S., Brozos, C., Fthenakis, G.C. Effects of drying-off procedure of ewes’ udder, with intramammary antibiotic administration, in subsequent mammary infection and development of mastitis. J. Hell. Vet. Med. Soc., 2012; 63:273-282.
- Jablonski, L.M., Bohach, G.A. Staphylococcus aureus. In: Food Microbiology: fundamentals and frontiers. Doyle, M.P., Beuchat, L.R., Montville, T.D. (eds.). ASM Press, Washington, D.C., USA, pp. 353-375.
- Boerema, J.A., Clemens, R., Brightwell, G. Evaluation of molecular methods to determine enterotoxigenic status and molecular genotype of bovine, ovine, human and food isolates of Staphylococcus aureus. Int. J. Food Microbiol., 2006; 107:192–201.
- Normanno, G., Corrente, M., Salandra, G.l., Dambrosio, A., Quaglia, N.C., Parisi, A., Santagada, G., Firinu, A., Crisetti, E., Celano, G.V. Methicillin resistant Staphylococcus aureus (MRSA) in foods of animal origin product in Italy. Int. J. Food Microbiol., 2007. 117:219-222.
- David, M.Z., Daum, R.S. Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin. Microbiol. Rev., 2010; 23: 616-687.
- Kumar, R., Yadav, B.R., Singh, R.S. Antibiotic resistance and pathogenicity factors in Staphylococcus aureus isolated from mastitic Sahiwal cattle. J. Bio. sci., 2011; 36:175–188.
- Vanderhaeghen, W., Cerpentier, T., Adriaensen, C., Vicca, J., Hermans, K., Butaye, P. Methicillin-resistant Staphylococcus aureus (MRSA) ST398 associated with clinical and subclinical mastitis in Belgian cows. Vet. Microbiol., 2010; 144:166-171.
- Lee, J.H., Methicillin (oxacillin)-resistant Staphylococcus aureus strains isolated from major food animals and their potential transmission to humans. Appl. Environ. Microbiol., 2003; 69: 6489-6494.
- Juhasz-Kaszanyitzky, E., Janosi, S., Somogyi, P. MRSA transmission between cows and humans. Emerg. Infect. Dis., 2007; 13: 630-632.
- Kluytmans, J.A. Methicillin-resistant Staphylococcus aureus in food products: cause for concern or case for complacency. Clin. Microbiol. Infect., 2010 ; 16:11-15.
- Unal, N., Askar, S., Macun, H.C., Sakarya, F., Altun, B., Yýldýrým, M. Panton–Valentine leukocidin and some exotoxins of Staphylococcus aureus and antimicrobial susceptibility profiles of staphylococci isolated from milks of small ruminants. Trop. Anim. Health. Prod., 2012; 44:573–579.
- Wilson, K Preparation of genomic DNA from bacteria. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, (eds) Current Protocols in Molecular Biology, 1987; pp. 2.4.1–2.4.5. John Wiley & Sons, New York.
- Braoios, A., Fluminhan J. A., Pizzolitto, A.C., Multiplex PCR use for Staphylococcus aureus identification and oxacillin and mupirocin resistance evaluation. Journal of Basic and Applied Pharmaceutical Sciences, 2009, 30(3):303-307
- CLSI, 2012. Performance standards for antimicrobial susceptibility test. M100-S22, CLSI, 32(3).
- Sudhan, N.A., Singh, R., Singh, M., Soodan, J.S. Studies on prevalence, etiology and diagnosis of subclinical mastitis among crossbred cows. Indian J. Anim. Res., 2005; 39: 127-130.
- Shrestha, S., Bindari, Y.R. Prevalence of subclinical mastitis among dairy cattle in Bhaktapur district, Nepal. Inter. J. Agri. Biosci., 2012; 1: 16-19.
- Rana, C.B., Prevalence of subclinical mastitis in bovines of Pokhara valley. BVSc and AH Internship Report, 2009; IAAS, Rampur.
- Abera, M., Elias, B., Aragaw, K., Denberga, Y., Amenu, K., Sheferaw, D. Major causes of mastitis and associated risk factors in smallholder dairy cows in Shashemene, southern Ethiopia. Afric. J. Agric. Res., 2012; 7: 3513-3518.
- Joshi, L.R., Tiwari, A., Devkota, S.P., Khatiwada, S., Paudyal, S., Pande, K.R. Prevalence of methicillin-resistant Staphylococcus aureus (MRSA) in dairy farms of Pokhara, Nepal. Inter. J. Vet. Sci., 2014; 3: 87-90.
- Mirzaei, H., Farhoudi, H., Tavassoli, H., Farajli, M., Monadi, A. Presence and antimicrobial susceptibility of methicillin resistant Staphylococcus aureus in raw and pasteurized milk and ice cream in Tabriz by culture and PCR techniques. Afr. J. Microbiol. Res., 2012; 6: 6224-6229.
- Alian, F., Rahimi, E., Shakerian, A., Momtaz, H., Riahi, M., Momeni, M. Antimicrobial resistance of Staphylococcus aureus isolated from bovine, sheep and goat raw milk. Global Veterinaria., 2012; 8:111-114.
- Nunes, S.F., Bexiga, R., Cavaco, L.M., Vilela, C.L. Technical note: Antimicrobial susceptibility of Portuguese isolates of Staphylococcus aureus and Staphylococcus epidermidis in subclinical bovine mastitis. J. Dairy Sci., 2007; 90:3242-3246.
- Virgin, J.E., Van Slyke, T.M., Lombard, J.E., Zadoks, R.N. Short communication: methicillin-resistant Staphylococcus aureus detection in US bulk tank milk. J. Dairy Sci., 2009; 92: 4988-4991.
- Coelho, S.M.O., Reinoso, E., Pereira, I.A., Soares, L.C., Demo, M., Bogni, C., Souza, M.M.S. Virulence factors and antimicrobial resistance of Staphylococcus aureus isolated from bovine mastitis in Rio de Janeiro. Pesq. Vet. Bras., 2009; 29: 369-374.
- Abebe, M., Daniel, A., Yimtubezinash, W., Genene, T. Identification and antimicrobial susceptibility of Staphylococcus aureus isolated from milk samples of dairy cows and nasal swabs of farm workers in selected dairy farms around Addis Ababa, Ethiopia. Afr. J. Microbiol. Res., 2013; 7: 3501-3510.
- Abera, M., Demie, B., Aragaw, K., Regassa, F., Regassa, A. Isolation and identification of Staphylococcus aureus from bovine mastitic milk and their drug resistance patterns in Adama town, Ethiopia. J. Vet. Med. Ani. Health, 2013; 2 :29-34.
- El-Jakee, J., Nagwa, A.S., Bakry, M., Zouelfakar, S.A., Elgabry, E., El-Said, W.A.G., Characteristics of Staphylococcus aureus strains isolated from human and animal sources. American-Eurasian J. Agric. Environ. Sci., 2008; 4: 221–229.
- Edward, M., Anna, K., Michal, K., Henryka, L., Krystyna, K. Antimicrobial susceptibility of staphylococci isolated from mastitic cows. Bull. Vet. Inst. Pulawy, 2002; 289-294.

