ISSN: 0973-7510
E-ISSN: 2581-690X
This study aims to present knowledge on therapeutic potential bacteriocin producing bacterial strains. Samples (n=35) from diverse Ernakulam sources yielded 26 lactic acid bacteria (LAB) isolates on De Man Rogosa Sharpe (MRS) medium. Among the isolates potent antimicrobial activity observed against known pathogens in one of the isolates from goat milk sample number 3, labelled as GO3. Microscopic and biochemical test revealed the isolate is Gram-positive rod, non-sporulating and resilience to pH and bile salts, with adaptability to gastric enzymes and surfactants. Genetic and physiological traits of the positive strains were confirmd by 16S rRNA technique. Blast analysis revealed that sequence of the strain GO3 from goat milk identified as Lactobacillus casei, exhibited antibiotic sensitivity, auto-aggregation, co-aggregation, and antibiofilm assays highlighted inhibitory properties as a promising therapeutic potential probiotic bacterium. LAB isolates can be further investigated at the genetic level to enhance their probiotic characteristics such as resistance and cellular adhesion. Additionally, the identification of gene responsible for expressing bacteriocins can be conducted on these isolates.
Bacteriocin, Lactic Acid Bacteria (LAB), Probiotics, Antimicrobial Activity, Therapeutic Potential
Lactobacilli play a crucial role in fermented dairy products and contribute to the therapeutic aspects of human health,1 probiotic can be found in dairy and non-dairy products. Lactic acid bacteria (LAB) are found in nature in large quantities and have potential role in medicine,2 but widely used as bio preservatives in food industries and adjuvants.3,4 Bacteriocins are antimicrobial proteins that Lactobacilli are known to produce, which are having antimicrobial property. The most prevalent genus of bacteria found in probiotics is Lactobacillus known as probiotics, confer health benefits to their host. Lactobacilli increase the acidity of the gastrointestinal system; it is believed that acidic environment will boost nutritional absorption and inhibit the growth of dangerous microorganisms. Additionally, lactobacilli may modify immune system function and reduce inflammation by secreting anti-inflammatory cytokines. Lactobacilli come in a wide variety of forms, and each has a special set of traits. Show therapeutic promise against genetic and metabolic diseases,5 gastrointestinal dysbiosis,6 and neurodegenerative disorders.7 Clinical trials in humans and mice demonstrate efficacy in managing various conditions, from irritable bowel symptoms to colorectal cancer, breast cancer and bladder cancer.6,7 Isolation, characterization and molecular identification of novel LAB stain has some advantage. It reveals their characteristics properties and help to obtain a promising functional LAB.8 Due to the ability of lactobacilli to thrive in diverse environment and complex microbial ecosystem, showcase their versatility and potential role in various application. Investigating lantibiotics or bacteriocin derived from these bacterial strains will yield valuable insights into the utilization of milk and dairy products as effective mediums for multistrain probiotic cultures. Additionally, it will shed light on the therapeutic applications of lantibiotics derived from milk, further highlighting its potential as a source of beneficial compounds for medical purposes.
Collection of Samples and Isolation of Lactic Acid Bacteria (LAB)
Thirty-five samples from different sources, rich in LAB such as Milk, dairy, fermented foods, and cattle dung were collected from Ernakulam District in Kerala and screened for Lactobacillus sp. The MRS medium (HiMedia-India) was used for isolation and culturing of Lactobacillus. Serial dilution and streak plate methods were used for isolation, and pure cultures were maintained in MRS broth (HiMedia-India) and stored in 20% glycerol (HiMedia-India) at -20°C for further investigation with slight modification.9
Screening of LAB Isolates for Antimicrobial Potential
Cell-free supernatant was prepared by selecting an isolated colony from the MRS agar plate culture, using 5ml MRS broth solution culture was incubated at 37°C for 24h for sub culture. Then 10 μl LAB culture was inoculated in 150 ml MRS broth and incubated at 37°C for 3 days. To get culture filtrate, broth culture centrifuged at 10,000 rpm for 20 min, at 4°C.10-12
Well diffusion method
Used for screening of LAB isolates against Bacillus subtilis, Escherichia coli, Klebsiella pneumonia, Salmonella typhi, Staphylococcus aureus, and Pseudomonas aeruginosa method given by Qin et al.13 and the width of the clear zone around producer colonies was measured after incubation on MHA (Himedia-India), recording positive inhibition if it was 2 mm or larger, with Tetracycline (15 µg as standard) because of its broad-spectrum antibiotic action.
Characterisation and identification of isolated LAB strains
Morphological Characterization Techniques
The initial bacterial identification involves morphological assessment, including Gram staining, motility, and endospore tests.14
Gram staining and cell morphology
Isolates were examined under a light microscope, and cell morphology is observed through Scanning Electron Microscope (ZEISS, Gemini SEM) at an accelerating voltage of 1.0 kv analysis. SEM involves fixing, dehydrating, and sputter-coating with gold bacterial cultures before observation.
Endospore and motility test
Schaeffer and Fulton staining technique utilized for spore and motility observed through Hanging drop method using light microscope.
The catalase test
Determines a positive response through effervescence after adding 3% hydrogen peroxide.
Acid tolerance
Preliminary selection of acid-tolerant lactobacilli using a rapid method was determined according to slightly modified methods15 to simulate gastric conditions. Lactobacilli were cultured into pH-adjusted MRS broth (2, 3 and 7), and OD monitored at 520 nm after 6 and 24h. Surviving indices, with a percentage equal to or greater than 50%, determined isolates’ viability
Bile salt tolerance
A modified method given16 is used for estimation of bile salt tolerance in a similar method of acid tolerance. An aliquot of 1 ml of the 6hour old culture inoculated in to 100 ml MRS broth with 0.2 or 0.4% bile salts, and 520 nm monitoring revealed surviving indices.
Growth at different temperatures
Bacterial strains cell density of 108 cfu/ml were inoculated to two different tubes of 5 ml MRS broth. Tubes were incubated overnight in a shaking incubator at various temperatures of 37°C and 45°C The turbidity of the growth of the strains at varying temperatures was compared with the control tube which was incubated at 37°C for 24 h by measuring in a colorimeter at 520 nm after overnight incubation at 37°C.17
NaCl tolerance
5 ml MRS broth were adjusted with different concentrations (0, 2, 3, 4 and 6.5%) of NaCl, each test tube was incubated with LAB strains 1.5 × 108 cfu/ml at 37°C for 24 h. Their growth was determined by observing their turbidity at 520 nm.18
Sensitivity to gastrointestinal enzymes
2 μl cell suspension (24 h growth) of the LAB isolate (108 cfu/ml) was swabbed on MRS agar. Disc was impregnated in prepared enzymatic solutions (Trypsin, lysozyme, α-amylase and proteinase-K) and placed on MRS agar plate then incubated at 37°C for 2 h19 and Zone of inhibition surrounding the disc were observed.
Surfactant sensitivity
Tested with 1% Tween 20, SDS, and EDTA. Solutions were mixed with LAB suspensions (108 cfu/ml), and OD at 520 nm was monitored after 60 min. Controls had cell suspensions without surfactants.20
Biochemical characterization methods
Biochemical characterization utilized the KB002 HiassortedTM Biochemical Test kit (Hi Media) was used for identification of LAB isolate. The kit contained a combination of 12 test, included 7 conventional biochemical tests (citrate utilization, lysine utilization, ornithole utilization, urease, phenylalanine deamination, nitrate reduction and H2S production and 5 carbohydrates tests. Interpretation was made based on the kit’s chart.
Specific biochemical tests
Methyl Red (MR), Voges-Proskauer (VP), and Kovac’s (Indole) tests were performed.
Antibiofilm test
Staphylococcus aureus colony was used for antibiofilm testing and aseptically transferred into 10 ml of nutrient broth, capped, and incubated overnight at 37°C. Biofilm inhibition was studied in a 96-well plate method with varied volumes (20-100 μl) of bacterial suspension along with 100 μl of nutrient broth. After 2 days at 37°C, 95% ethanol was added and incubated for 15 minutes, and spectrophotometric readings at 570 nm were recorded for the reaction mixture.21
Antibiotic susceptibility test
LAB isolates were tested for 9 antibiotics using the agar overlay diffusion method as per National Committee for Clinical Laboratory Standards.22 Muller Hinton Agar medium was used for the study and result were expressed according to cut off levels proposed.23
Microbial adhesion to hydrocarbons
Microbial adhesion to hydrocarbons (MATH) slightly modified method of Rijnaarts et al.24 used culturing cells in MRS broth at 37°C for 18 h. After centrifugation, cells were washed with PBS, resuspended in KNO3 (pH 6.2), and adjusted to 108 cfu/ml. Following pre-incubation, a two-phase system was created, vortexed, and incubated. A1 absorbance at 600 nm determined microbial adhesion percentage using the formula.
MATH (%) = [ 1 – A1/A0 ] × 100 …(1)
Auto-aggregation and co-aggregation method
Auto-aggregation was evaluated by25 inoculating 20 μl of LAB culture into 150 ml MRS broth, incubating at 37°C for 24 h. After harvesting the cells (7000 g, 10 min, 20°C), they were washed, resuspended in sterile physiological water, and diluted to OD~0.3 at 660 nm. A 2 ml sterile plastic cuvette was filled with 1 ml of the cell suspension, and OD at 660 nm was recorded for 60 min using a microplate Reader (Cyber Elisa RO1, Cyberlab USA). Auto-aggregation degree was calculated using the provided equation.
Auto aggregation (%) = [1-At/A0] × 100 …(2)
Where At represents the absorbance at time t=60 min and A0 the absorbance at t=0.
The co-aggregation activity
Selected LAB was assessed similarly to auto-aggregation.25 Specifically, 20 μl of each LAB culture and E. coli culture were inoculated into 150 ml MRS broth and Nutrient broth, respectively, and incubated at 37°C for 24 h. After incubation, 1 ml of LAB culture cell suspension was combined with 1 ml of E. coli culture, transferred to a 2 ml sterile cuvette, and OD was measured at 660 nm over 60 min using a microplate Reader (Cyber Elisa RO1, Cyberlab USA). Co-aggregation (%) was calculated using the formula:
Co-aggregation(%) = [ (ODx + ODy /2) – 0(x+y) / ( ODx + ODy / 2) ] × 100 …(3)
Where x and y represent each of the two strains in the control tubes, and (x + y) the mixture.
Autolytic assay
The procedure for preparing cell suspensions for autolysis was identical to that used in auto aggregation assays. Absorbance measurements were taken at 3 and 5 hours after mixing the cell suspensions. The percentage of autolytic activity was determined using the following formula:
Autolytic Assay= [1-At/A0] × 100 …(4)
Where At represents the optical density after mixing in MRS at time t = 3 or 5 h and A0 the optical density after mixing at t =0.
Molecular characterization
Traditional microbiological methods include morphological, biochemical, and physiological techniques, while 16S rRNA sequencing is a reliable method for bacterial identification. Prokaryotes have 70S ribosomes with 30S subunits containing conserved 16S rRNA for species-specific identification. This approach assesses prokaryotic diversity and phylogenetic relationships in various sources, including environmental and fermented samples.26,27
Extraction of bacterial genomic DNA
Genomic DNA extraction was carried out using chloroform: isoamyl alcohol method. Selected LAB isolated was cultured in Nutrient broth overnight (12-16 hours). 1ml of overnight culture was centrifuged. The pellet was resuspended in 1 ml 0.85% (w/v) NaCl, treated with lysis buffer and proteinase-K, followed by incubation at 65°C. Chloroform: Isoamyl alcohol extraction, was repeated and the aqueous phase phase which in turn mixed with sodium acetate and Isopropanol. After centrifugation, the DNA pellet was rinsed, desiccated, re-suspended in 100 μl TE buffer and refrigerated.
Identification of potential strains by 16s rRNA or 16s rDNA
The positive strains were confirmed by 16S ribosomal RNA (rRNA) analysis. PCR amplification was performed using the 16S rDNA. Two primers namely forward primer 16F (5’-AGA GTTTGATCCTGG-3’) and reverse primer 16R1492 (5’-ACCTTGTTACGACTT-3’) from Thermoscientific, USA. PCR mix: 0.5 μl of each primer, 13 μl Master Mix (Takara Emerald Amp GT PCR Master Mix), 9 μl dd.H2O, and 2 μl DNA Template. Amplification occurred in a SimpliAmp thermocycler (94°C, 2 min; 35 cycles of 94°C/45 s, 55°C/60 s, 72°C/60 s; final extension at 72°C/10 min). PCR products were electrophoresed in a 1.5% Agarose gel with Ethidium bromide. Visualization used a Gel documentation system after electrophoresis.
PCR product purification
The PCR product was purified using a commercial PCR clean-up kit (GenEluteTM PCR Clean-Up Kit Sigma) according to the manufacturer’s instructions.
DNA sequencing and identification of bacterial strains
DNA sequencing and bacterial strain identification involve determining the nucleotide sequence of a microorganism’s DNA.
Phylogenetic analysis
The Maximum Likelihood method and Tamura-Nei model were employed to infer the evolutionary history. The tree with the highest log-likelihood (-2090.79) is displayed, indicating the percentage of trees supporting taxa clustering. Initial trees were generated using Neighbor-Join and BioNJ algorithms, selecting the topology with superior log likelihood. Site proportions with unambiguous bases in each clade were shown. Analyses were performed in MEGA X.
Collection and isolation of samples
In this study, 35 samples were collected from different sources in Ernakulam District, Kerala, including milk, dairy products, fermented foods, and cattle dung. These samples were plated on MRS agar plates, and after incubation, 26 lactic acid bacteria (LAB) isolates were obtained. The LAB isolates were categorized as CO: Cow milk, BU: Buffalo milk, GO: Goat milk, CR: Curd, CH: Cheese, DS: Dosa, ID: Idli, CG: Cow dung, and BG: Buffalo dung based on their respective sources of origin (Table 1, 2).
Table (1):
Colony Forming Unit (CFU) values of samples
| Source | Sample | Colony Forming Unit |
|---|---|---|
| Buffalo Milk (4) | sample1 | 2.5 x 10 |
| sample2 | 0 | |
| sample3 | 2.1 x 103 | |
| sample4 | 2.4 x 103 | |
| Cow Milk (5) | sample1 | 6.0 x 102 |
| sample2 | 2.2 x 103 | |
| sample3 | 0 | |
| sample4 | 2.4 x 103 | |
| sample5 | 1.8 x 103 | |
| Goat Milk (5) | sample1 | 0 |
| sample2 | 2.6 x 103 | |
| sample3 | 5.0 x 102 | |
| sample4 | 0 | |
| sample5 | 2.3 x 102 | |
| Cheese (4) | sample1 | 7.8 x 102 |
| sample2 | 8.1 x 103 | |
| sample3 | 3.5 x 104 | |
| sample4 | 2.0 x 102 | |
| Curd (7) | sample1 | 3.1 x 104 |
| sample2 | 3.5 x 106 | |
| sample3 | 3.9 x 106 | |
| sample4 | 1.8 x 108 | |
| sample5 | 2.0 x 1070 | |
| sample6 | 0 | |
| sample7 | 2.6 x 107 | |
| Dosa (3) | sample1 | 2.4 x 102 |
| sample2 | 0 | |
| sample3 | 1.8 x 102 | |
| Idli (2) | sample1 | 1.0 x 102 |
| sample2 | 0 | |
| Cow Dung (3) | sample1 | 0 |
| sample2 | 2.8 x 101 | |
| sample3 | 0 | |
| Buffalo Dung (2) | sample1 | 0 |
| sample2 | 3.6 x 101 |
Table (2):
Isolation and screening of isolated Lactic Acid Bacteria (LAB)
S.N. |
Sources |
No. of samples |
No. of isolates |
Positive Bacteria |
|---|---|---|---|---|
1 |
Buffalo Milk (BU) |
4 |
3 |
1 |
2 |
Cowmilk (CO) |
5 |
4 |
2 |
3 |
Goat Milk(GO) |
5 |
4 |
3 |
4 |
Cheese(CH) |
4 |
3 |
2 |
5 |
Curd (CR) |
7 |
7 |
4 |
6 |
Dosa (DS) |
3 |
2 |
1 |
7 |
Idli (ID) |
2 |
1 |
0 |
8 |
Cow Dung (CG) |
3 |
1 |
0 |
9 |
Buffalo Dung (BG) |
2 |
1 |
1 |
Total |
35 |
26 |
14 |
Screening of lactic acid bacteria for antimicrobial potential
Competitive action of LAB for nutrients and production of antimicrobial compounds like organic acid, hydrogen peroxide, bacteriocins antifungal peptides responsible for their antimicrobial properties. Lactobacillus isolates with antimicrobial activity, regarded as positive. Out of the 26 LAB isolates, only 14 strains exhibited antimicrobial activity against six prevalent foodborne pathogens, namely Bacillus subtilis, Escherichia coli, Klebsiella pneumonia, Salmonella typhi, Staphylococcus aureus, and Pseudomonas aeruginosa. Isolate GO3, showed prompting antimicrobial property, selected for further investigation (Table 3).
Table (3):
Degree of zone of inhibition of LAB isolates
| Indicator organisms | Zone of inhibition | Strains |
|---|---|---|
| Escherichia coli, Pseudomonas aeruginosa, Staphylococcus aureus, Salmonella typhi, Klebsiella pneumoniae and Bacillussubtilis | +++ | BU1, GO3, CR5 |
| ++ | GO1, GO2, CO3, BG1 | |
| + | DS2, CR4, CR7, DS2, CG1, BU2, CH3 |
(+++: Inhibition area per well is of >8.0 % of the petri dish; ++: Inhibition area per well is of 3.0-8.0% of the petri dish; and +: Inhibition area per well is of 0.1-3.0% of petri dish)
Three isolates, namely BU1, GO3, and CR5, displayed maximum zones of inhibition, exceeding 8 mm. Notably, GO3, isolated from goat milk, demonstrated the highest antimicrobial potential (Table 4).
Table (4):
Diameter of Zone of inhibition in mm of potent LAB isolates
| S.N. | Isolate | Pathogenic Bacteria | |||||
|---|---|---|---|---|---|---|---|
| Bacillus subtilis | Escherichia coli | Pseudomonas aeruginosa | Klebsiella pneumoniae | Staphylococcus aureus | Salmonella typhi | ||
| 1 | BU1 | 13 ± 0.05 | 26 ± 0.02 | 12 ± 0.03 | 21 ± 0.06 | 12 ± 0.05 | 23 ± 0.02 |
| 2 | GO3 | 21 ± 0.08 | 28 ± 0.02 | 20 ± 0.05 | 16 ± 0.01 | 20 ± 0.03 | 28 ± 0.06 |
| 3 | CR5 | 20 ± 0.04 | 27 ± 0.02 | 16 ± 0.04 | 18 ± 0.05 | 17 ± 0.04 | 21 ± 0.02 |
| 4 | Tet | 25 ± 0.04 | 27 ± 0.06 | 16 ± 0.01 | 15 ± 0.02 | 19 ± 0.08 | 26 ± 0.05 |
Characterization and identification of isolated strains
Among the isolates GO3 showed significant anti-bacterial activity and, hence, was selected for further tests.
Morphological characterization
The morphological characterization involves the examination of cell morphology using gram staining, endospore staining and scanning electron microscopy (SEM). Gram staining revealed GO3, Gram-positive in nature, staining with Schaeffer and Fulton’s spore stain confirmed non-sporulation. Scanning Electron Microscopic (ZEISS GeminiSEM) analysis at 10.00KX, 25.00KX, 50.00KX, and 100.00KX magnifications revealed rod-shaped cells growing in pairs. Cultivated in MRS broth for 16 hours, the cells measured 1.2 µm to 1.9 µm in length and 350 nm to 450 nm in width (Table 5a).
Table (5a):
Morphological characteristics of GO3 Isolate
S.N. |
Morphological tests |
Isolate GO3 |
|---|---|---|
1 |
Gram staining |
Gram-positive |
2 |
Endospore test |
– |
3 |
Cell arrangements (SEM) |
single or pairs |
4 |
Cell Shape (SEM) |
Rods |
5 |
Cell size (SEM) |
Length: 1.2 µm-1.9 µm width: 350 nm-450 nm |
Additional tests of selected LAB strains
Performed catalase test and motility test, GO3 displayed catalase positivity, releasing oxygen effervescence with H2O2. Motility was not observed under a light microscope, indicating non-motility (Table 5b).
Table (5b):
Additional tests of selected LAB strains
S.N. |
Tests |
Isolate GO3 |
|---|---|---|
1 |
Catalase test |
+ |
2 |
Motility test |
– |
Physiological characterisation
Acid tolerance
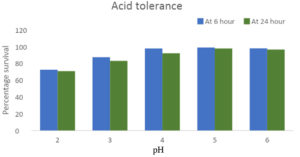
Under pH 2 conditions, after a 6-hour incubation, the survival of GO3 was determined to be 72.5%, and after 24 hours, it decreased to 70.8%. At pH 3, the survival percentage after 6 hours was 87.5%, and after 24 hours, it decreased to 83.5%. With an increase in pH to 6, the survival percentage at both 6 hours and 24 hours was higher compared to that at pH 2.0. Based on the classification criteria, strain GO3 is categorised as having ‘good tolerance’ to the tested pH conditions (Figure 1).
Figure 1. Survival percentage of GO3 isolate after 6h and 24h of incubation in pH 2-6 medium respectively
Bile salt tolerance
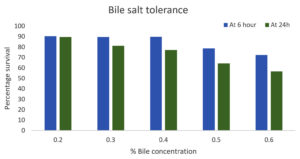
LAB isolates were cultured in MRS broth with 0.2%, 0.3%, 0.4%, 0.5%, and 0.6% bile salt (BS) concentrations, with a BS-free LAB culture as the control. At 0.4% BS, GO3’s survival was 89.8% at 6 hours, decreasing to 77.2% at 24 hours. For 0.2% BS, survival was 90.37% at 6 hours, decreasing to 89.65% at 24 hours. According to classification criteria, the isolate showed ‘good tolerance.’ The decreasing survival at higher BS concentrations indicated intolerance and reduced viability. With increased bile salt concentration, the survival percentage decreased after 24 hours, suggesting significant survival linked to bile salt hydrolases (BSH) in LAB isolates (Figure 2).
Growth at different temperatures
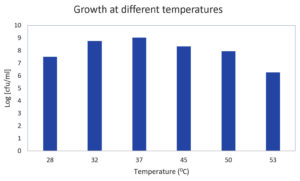
The chosen LAB strains (at a concentration of 108 cfu/ml) were introduced into 5 ml MRS broth tubes and incubated for 24 hours at various temperatures. Growth was assessed in terms of log CFU/ml. At 37°C, GO3 exhibited a peak growth of 9.03, and as the temperature rise to 53°C, the growth decreased to 6.25 (Figure 3). As per McFarland’s standard where OD 0.5 corresponds to 1.5 × 108 cfu/ml. All the calculations are made accordingly, hence measurement expressed in CFU/ml.
Growth at different NaCl concentrations
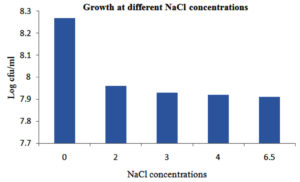
The chosen LAB strain, GO3, was cultivated in MRS broth with varying NaCl concentrations (0%, 2%, 3%, 4%, and 6.5%) for 24 hours. The observed growth (log cfu/ml) at the respective NaCl concentrations was 8.27, 7.96, 7.93, 7.92, and 7.91. As per McFarland’s standard where OD 0.5 corresponds to 1.5 × 108 cfu/ml. All the calculations are made accordingly, hence measurement expressed in cfu/ml (Figure 4).
Sensitivity to gastrointestinal enzymes
The impact of various gastrointestinal enzymes (trypsin, lysozyme, α-amylase, and proteinase-K) on the growth of LAB strains was evaluated. The growth of the GO3 isolate was unaffected by trypsin, α-amylase, and proteinase-K, while it exhibited slight sensitivity to lysozyme (Table 6).
Table (6):
Sensitivity against Digestive Enzymes
S.N. |
Gastrointestinal enzymes |
GO3 isolate |
|---|---|---|
1 |
Trypsin |
– |
2 |
Lysozyme |
+ |
3 |
α-amylase |
– |
4 |
Proteinase-K |
– |
Sensitivity to surfactants
When LAB strains (inoculum ~ 108 cfu/ml) were subjected to various surfactants for 60 minutes, the growth (log cfu/ml) of both strains was most impacted in the presence of SDS, decreasing to 7.74. Conversely, the growth increased to 8.24 and 8.10 in the presence of EDTA and Tween 20, respectively (Table 7).
Table (7):
Growth of GO3 in the presence of surfactants
Surfactants |
Growth cfu/ml |
|---|---|
EDTA |
8.24 |
SDS |
7.74 |
TWEEN 20 |
8.10 |
Biochemical characterization
Carbohydrate fermentation tests
In the case of the GO3 strain, the carbohydrate fermentation test demonstrated the ability to ferment Lactose, Xylose, Maltose, Dextrose, Galactose, L-arabinose, Mannose, and α-Methyl-D-mannoside. However, GO3 did not exhibit fermentation for Fructose, Raffinose, Trehalose, Melibiose, Sucrose, Inulin, Sodium gluconate, Glycerol, Salicin, Dulcitol, Inositol, Sorbitol, Mannitol, Adonitol, Arabitol, Erythritol, α-Methyl-D-glucoside, Rhamnose, Cellobiose, Melezitose, Xylitol, ONPG, Esculin hydrolysis, D-Arabinose, Citrate utilization, Malonate utilization, and Sorbose (Table 8).
Table (8):
Results of Carbohydrate Fermentation Tests
S.N. |
Tests |
Result |
|---|---|---|
1 |
Lactose |
+ |
2 |
Xylose |
+ |
3 |
Maltose |
+ |
4 |
Fructose |
– |
5 |
Dextrose |
+ |
6 |
Galactose |
+ |
7 |
Raffinose |
– |
8 |
Trehalose |
– |
9 |
Melibiose |
– |
10 |
Sucrose |
– |
11 |
L-Arabinose |
+ |
12 |
Mannose |
+ |
13 |
Inulin |
– |
14 |
Sodium gluconate |
– |
15 |
Glycerol |
– |
16 |
Salicin |
– |
17 |
Dulcitol |
– |
18 |
Inositol |
– |
19 |
Sorbitol |
– |
20 |
Mannitol |
– |
21 |
Adonitol |
– |
22 |
Arabitol |
– |
23 |
Erythritol |
– |
24 |
α-Methyl-D-glucoside |
– |
25 |
Rhamnose |
– |
26 |
Cellobiose |
– |
27 |
Melezitose |
– |
28 |
α-methyl-D-mannoside |
+ |
29 |
Xylitol |
– |
30 |
ONPG |
– |
31 |
Esculin hydrolysis |
– |
32 |
D-Arabinose |
– |
33 |
Citrate utilization |
– |
34 |
Malonateutilization |
– |
35 |
Sorbose |
– |
Biochemical tests
In the case of the GO3 strain, positive results were obtained for Phenylalanine Deamination, and it demonstrated the ability to ferment glucose (Table 9).
Table (9):
Biochemical Test Results
S.N. |
Biochemical Tests |
Results |
|---|---|---|
1 |
Citrate utilization |
– |
2 |
LysineUtilization |
– |
3 |
OrnithineUtilization |
– |
4 |
Urease |
– |
5 |
PhenylalanineDeamination |
+ |
6 |
NitrateReduction |
– |
7 |
H2S Production |
– |
8 |
Glucose |
+ |
9 |
Adonitol |
– |
10 |
Lactose |
– |
11 |
Arabinose |
– |
12 |
Sorbitol |
– |
Specific biochemical tests
In this study, isolates were subjected to Methyl Red (MR), Voges Proskauer (VP), and Indole tests, these are mainly used to determine the metabolic activity of Lactobacillus. The GO3 strain exhibited a positive reaction for the Indole test and negative reactions for both the Methyl Red and Voges Proskauer tests (Table 10).
Table (10):
Specific Biochemical Tests
S.N. |
Specific biochemical tests |
GO3 isolate |
|---|---|---|
1 |
Methyl Red Test |
– |
2 |
Voges Proskauer Test |
– |
3 |
Indole Test |
+ |
Antibiofilm test
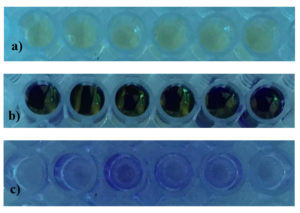
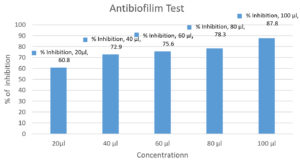
GO3’s antibiofilm efficacy was assessed using 100 μl Staphylococcus aureus suspension, percentage of inhibition become high when concentration of GO3 increased (Figure 5 & 6).
Figure 5. a) Microtitre plate before incubation, b) Microtitre plate on the addition of 1% Crystal Violet, c) Microtitre plate after washing with 95% ethanol
Figure 6. Inhibition of biofilm formation by different concentrations of Isolate GO3 against Staphylococcus aureus
Antibiotic susceptibility test
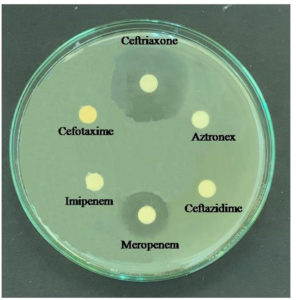
Selected LAB isolate was tested for susceptibility to six antibiotics—Aztronex, Ceftazidime, Ceftriaxone, Cefotaxime, Imipenem, and Meropenem—using the agar overlay diffusion method as per the National Committee for Clinical Laboratory Standards. GO3 exhibited sensitivity to Ceftriaxone and Meropenem but remained resistant to other antibiotics (Figure 7).
Microbial adhesion to hydrocarbons assay (MATH)
In this study, GO3 isolates were cultured with xylene, toluene, and n-hexadecane, non-polar solvents leveraging their hydrophobic properties to interact with microbial surfaces. The GO3 isolate exhibited microbial adhesion exceeding 40% in all three solvents (Table 11).
Table (11):
Microbial adhesion to hydrocarbons assay (MATH) (%) of GO3
Hydrocarbons |
MATH (%) |
|---|---|
Xylene |
52.4 |
Toluene |
54.3 |
n-Hexadecane |
60.8 |
Auto-aggregation and coaggregation
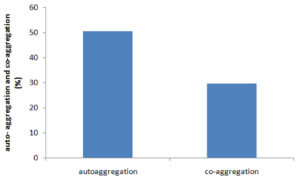
In this study, GO3 displayed auto-aggregation (50.6%) and co-aggregation (27.9% at 0h, 29.75% at 60 min), which ascertain efficiency of selected LAB (Figure 8).
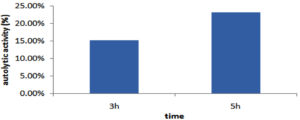
Autolytic assay
The autolytic rate of selected LAB measured in MRS medium after 3 and 5 hours, GO3 showed autolytic activity of 15.3% after 3 h and after 5 h, it increased to 23.2% respectively (Figure 9).
Molecular characterization
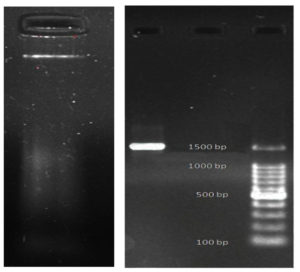
The DNA of strain GO3 was isolated for molecular identification through 16S rRNA sequencing. The nucleotide sequence was analyzed for sequence identify by BLAST in the GenBank of NCBI (Figure 10).
Phylogenetic and Evolutionary analysis by Maximum Likelihood method. BLAST analysis revealed a 100% sequence similarity between isolate GO3 and Lactobacillus casei strain NR115322.1 (Figure 11). and sequence length were 1500 bp. To obtain GenBank accession number, 16S rRNA sequence of strain were uploaded in to NCBI databases (on 24 March 2022), accession number is ON059683.
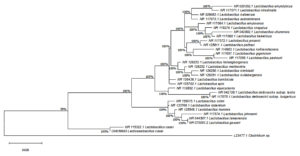
Evolutionary history was inferred by Maximum Likelihood method, using the Tamura-Nei model, The tree, with the highest log likelihood (-2090.79), displays taxonomic clustering percentages. Initial trees for the heuristic search were obtained with Neighbor-Join and BioNJ algorithms, using the Tamura-Nei model for pairwise distance estimation. The superior log likelihood topology was chosen. Proportions of sites with at least one unambiguous base in each descendant clade are indicated near internal nodes. This analysis involved 29 nucleotide sequences and 661 positions. Analyses were conducted in MEGAX (Figure 12).
The lactic acid bacteria are widely distributed in nature and abundantly found as indigenous microflora in raw milk, fermented dairy products like butter, cheese, curd, fermented milk, lassi whey, yoghurt and also in fermented drinks such as sorghum beer, cassava etc. In the present study, thirty-five samples were collected from Ernakulam District in Kerala. Samples were collected from different sources; milk, dairy, fermented foods and cattle dung. A total of 26 LAB isolates were grown on MRS agar plates as mentioned earlier,28 designated on the basis of their source of origin, then subjected to screening for antimicrobial potential of the strains. Evaluation of LAB isolated from different samples for biotherapeutic application not much explored previously to our knowledge. Although milk, dairy product, fermented food and cattle manure are easily available in Kerala. Detection of antimicrobial activity is one of the most important features of any viable probiotic strain; these properties were consequence of competition for nutrients and production of antimicrobial molecule such as lactic acid, acetic acid, bacteriocin, antifungal peptides and hydrogen peroxide.29 Among 26 LAB isolate only 14 strains showed antimicrobial activity against 6 common food borne pathogens, i.e., Bacillus subtilis, Escherichia coli, Klebsiella pneumonia, Salmonella typhi, Staphylococcus aureus and Pseudomonas aeruginosa by well diffusion method which is an established method to study antimicrobial action of LAB.9 Maximum zone of inhibition (≤ 8mm) were exhibited by 3 isolates: BU1, GO3, CR5. However, GO3 isolated from goat milk sample number 3, showed highest zone of inhibition against, Salmonella typhi (28 ± 0.06 mm), Escherichia coli (28 ± 0.02mm) and intermediate activity was recorded against Bacillus subtilis (21 ± 0.08), Pseudomonas aeruginosa (20 ± 0.05), Staphylococcus aureus (20 ± 0.03+) and Klebsiella pneumoniae(16 ± 0.01) This finding was conflict to Sharifi-Yazdi et al.30 investigation reported highest action against gram-positive bacteria than gram-negative bacteria. From the previous finding of Jose et al. and Setyawardani et al.,31,32 reported LAB strain have broad spectrum of anti-microbial action against both gram-positive and gram-negative bacteria. Among the isolate significant anti-bacterial activity showed GO3 selected for further tests. Morphological and physiological analyses of selected LAB unveiled Gram-positive nature, non-sporulating behaviour, and non-motile characteristics. Scanning electron microscopy illustrated its rod-shaped cell morphology and resilience to various conditions. Propitious feature of bacteriocin producing probiotic strain depends on the survival rate of acidic pH, bile salt tolerance etc. In this research viability of GO3 observed using different physiological parameters such as acid-bile salt tolerance, sensitivity to digestive enzyme and surfactance, growth at different temperature and NaCl. GO3 exhibited good survival category of acidic pH (2 and 3) similarly reported33 and bile salt tolerance (0.2% and 0.4%) previously mentioned33 without damaging viability of cell, live bacterial probiotics contribute to stimulate digestive enzyme synthesis, GO3 was not damaged in the presence of trypsin, α-amylase and proteinase-K, this finding was supported with Taoka et al.34 Different surfactants were tested with GO3, including SDS, Tween-20 and EDTA at 1% w/v concertation. The highest rate of inhibition was observed in SDS, while lowest in EDTA, this finding was contradictory to the Le et al.35 Bacterial physiology differs from one another, in order to identify metabolic ability of G03 performed carbohydrate and biochemical tests. Active probiotic strains have the ability to adhere intestinal mucus then compete and displace the adhesion of infectious pathogen.36 Selected LAB showed auto aggregation and co-aggregation property with pathogen, this activity indicate strain can produce antimicrobial agent to inhibit pathogen adhesion.37 Microbial adhesion to hydrocarbons (MATH) is helpful to study easily adhesion property of probiotic to prevent colonization of pathogen in gastrointestinal tract. In this GO3 isolate showed microbial adhesion exceeding 40%, probiotic strain typically requires a minimum of 40% MATH in all three solvents.38 Moreover, the CFS of selected strain exhibited active antibiofilm effect using Staphylococcus aureus previously investigated.39 In this study autolytic activity of the strain measured in MRS medium, autolysis property of GO3 found to be increasing while increasing temperature. This property of LAB can explored in food industry for texture and flavor development of the dairy product studies reported.37 Molecular identification of GO3 through 16S rRNA sequencing by BLAST in the GenBank of NCBI showed 100% similarity with Lactobacillus casei strain NR115322.1, 1500 bp sequence length and accession number is ON059683. Findings of the present study pointed out that the selected LAB from goat milk is efficient to develop as therapeutic agent or as probiotic component in food industry for flavor and texture improvement in future without any side effects.
Based on the findings of present study, Lactobacillus casei isolated from goat milk collected from the local region of Ernakulam-Kerala, showing prominent characteristic features of potent bacteriocin contain bacteria, which can be developed as a candidate for therapeutic purpose and industrial application in future. Therefore, Lactobacillus casei stored as a research tool to study and improve bacteriocin activity and therapeutic power along with molecular identification of gene for biotherapeutic application.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This article does not contain any studies on human participants or animals performed by any of the authors.
- Kajal A, Ankur G, Jagriti S. Isolation and identification of Lactobacilli bacteria from raw cow milk in local region of Agra. Int J Adv Res Biol Sci. 2017;4(11):98-102.
Crossref - Stavropoulou E, Bezirtzoglou E. Probiotics in medicine: a long debate. Front Immunol. 2020;11:2192.
Crossref - Asteri IA, Kittaki N, Tsakalidou E. The effect of wild lactic acid bacteria on the production of goat’s milk soft cheese. Int J Dairy Technol. 2010;63(2):234-242.
Crossref - Psoni L, Kotzamanidis C, Yiangou M, Tzanetakis N, Litopoulou-Tzanetaki E. Genotypic and phenotypic diversity of Lactococcus lactis isolates from Batzos, a Greek PDO raw goat milk cheese. Int J Food Microbiol. 2007;114(2):211-220.
Crossref - Bi M, Liu C, Wang Y, Liu SJ. Therapeutic Prospect of New Probiotics in Neurodegenerative Diseases. Microorganisms. 2023;11(6):1527.
Crossref - Harper A, Naghibi MM, Garcha D. The role of bacteria, probiotics and diet in irritable bowel syndrome. Foods. 2018;7(2):13.
Crossref - Rafter J. Probiotics and colon cancer. Best Pract Res Clin Gastroenterol. 2003;17(5):849-859.-colon cancer.
Crossref - Bin Masalam MS, Bahieldin A, Alharbi MG, et al. Isolation, molecular characterization and probiotic potential of lactic acid bacteria in Saudi raw and fermented milk. Evid Based Complement Alternat Med. 2018;2018:33.
Crossref - Veettil VN, Chitra AV. Probiotic Lactic Acid Bacteria from Goat’s Milk Potential Producer of Bacteriocin:Evidence from Liquid Chromatography-Mass Spectrometry. J Pure Appl Microbiol. 2022;16(1):305-317.
Crossref - Khochamit N, Siripornadulsil S, Sukon P, Siripornadulsil W. Antibacterial activity and genotypic-phenotypic characteristics of bacteriocin-producing Bacillus subtilis KKU213:potential as a probiotic strain. Microbiol Res. 2015;170:36-50.
Crossref - Xie J, Zhang R, Shang C, Guo Y. Isolation and characterization of a bacteriocin produced by an isolated Bacillus subtilis LFB112 that exhibits antimicrobial activity against domestic animal pathogens. Afr J Biotechnol. 2009;8(20).
- Mah JH, Kim KS, Park JH, Byun MW, Kim YB, Hwang HJ. Bacteriocin with a broad antimicrobial spectirum, produced by Bacillus sp. isolated from Kimchi. J Microbiol Biotechnol. 2001;11(4):577-584.
- Qin Y, Wang Y, He Y, et al. Characterization of subtilin L-Q11, a novel class I bacteriocin synthesized by Bacillus subtilis L-Q11 isolated from orchard soil. Front Microbiol. 2019;10:484.
Crossref - Balouiri M, Sadiki M, Ibnsouda SK. Methods for in vitro evaluating antimicrobial activity:a review. J Pharm Anal. 2016;6(2):71-79.
Crossref - Sieladie DV, Zambou NF, Kaktcham PM, Cresci A, Fonteh F. Probiotic properties of Lactobacilli strains isolated from raw cow milk in the western highlands of Cameroon. Innov Rom Food Biotechnol. 2011;9:12-28.
- Dora IA, Glenn RG. Cholesterol assimilation by lactic acid bacteria and bifidobacteria isolated from the human gut. Appl Environ Microbiol. 2002;68(9):4689-93.
Crossref - Fitriani VY, Suprapti B, Amin M. The characteristics of lactic acid bacteria isolated from fermented food as potential probiotics. J Basic Clin Physiol Pharmacol. 2021;32(4):743-749.
Crossref - Wu H, Shum TF, Chiou J. Characterization of the Probiotic Potential of Lactic Acid Bacteria Isolated from Kimchi, Yogurt, and Baby Feces in Hong Kong and Their Performance in Soymilk Fermentation. Microorganisms. 2021;9(12):2544.
Crossref - Mulaw G, Sisay Tessema T, Muleta D, Tesfaye A. In vitro evaluation of probiotic properties of lactic acid bacteria isolated from some traditionally fermented Ethiopian food products. Int J Microbiol. 2019;11.
Crossref - Neha J, Archana M, Vandana B. Screening, Characterization and In Vitro Evaluation of Probiotic Properties of Lactobacillus Strains. Asian J Pharm Clin Res. 2017;10(8):288.
Crossref - Usman RA, Taura DW, Bukar A. Antagonistic Activity of Partially Purified Bacteriocins Produced by Lactobacillus species Isolated from Nono (Fermented Milk). Nigerian Journal of Microbiology. 2022;36(1):6141-6150.
- Kiehlbauch JA, Hannett GE, Salfinger M, Archinal W, Monserrat C, Carlyn C. Use of the National Committee for Clinical Laboratory Standards guidelines for disk diffusion susceptibility testing in New York state laboratories. Journal of clinical microbiology. 2000, 38(9):3341-3348.
Crossref - Vlkova E, Rada V, Popelarova P, Trojanova I, Killer J. Antimicrobial susceptibility of bifidobacteria isolated from gastrointestinal tract of calves. Livestock Science. 2006;105(1-3):253-259.
Crossref - Rijnaarts HH, Norde W, Bouwer EJ, Lyklema J, Zehnder AJ. Bacterial adhesion under static and dynamic conditions. Appl Environ Microbiol. 1993;59(10):3255-3265.
Crossref - Basson A, Flemming LA, Chenia HY. Evaluation of adherence, hydrophobicity, aggregation, and biofilm development of Flavobacterium johnsoniae-like isolates. Microbial Ecology. 2008;55(1):1-14.
Crossref - Alziadi RE. Detection of genes encode for producing Bacteriocins in Lactobacillus genus isolated from local Iraqi dairy products. Muthanna Medical Journal. 2017;4(6):168-178.
- Tafvizi F, Tajabadi Ebrahimi M, Khojare L. Genotypic and phylogenic analysis of lactobacilli producing bacteriocin isolated from traditional dairy products and food. Journal of Fasa University of Medical Sciences. 2012;2(2):84-90.
- Reuben RC, Roy PC, Sarkar SL, Alam ARU, Jahid IK. Characterization and evaluation of lactic acid bacteria from indigenous raw milk for potential probiotic properties. J Dairy Sci. 2020;103(2):1223-1237.
Crossref - Grosu-Tudor SS, Stancu MM, Pelinescu D, Zamfir M. Characterization of some bacteriocins produced by lactic acid bacteria isolated from fermented foods. World J Microbiol Biotechnol. 2014;30(9):2459-2469.
Crossref - Sharifi-Yazdi MK, Davoodabadi A, Khesht Zarin HR, Tajabadi Ebrahimi M, Soltan Dallal MM. Characterisation and probiotic potential of lactic acid bacteria isolated from Iranian traditional yogurts. Ital J Anim Sci. 2017;16(2):185-188.
Crossref - Jose NM, Bunt CR, Hussain MA. Comparison of microbiological and probiotic characteristics of lactobacilli isolates from dairy food products and animal rumen contents. Microorganisms. 2015;3(2):198-212.
Crossref - Setyawardani T, Rahayu WP, Maheswari RR, Palupi NS. Antimicrobial activity and adhesion ability of indigenous lactic acid bacteria isolated from goat milk. Int Food Res J. 2014;21(3):959-964.
- Khushboo, Karnwal A, Malik T. Characterization and selection of probiotic lactic acid bacteria from different dietary sources for development of functional foods. Front Microbiol. 2023;14:1170725.
Crossref - Taoka Y, Maeda H, Jo JY, Sakata T. Influence of commercial probiotics on the digestive enzyme activities of Tilapia, Oreochromis niloticus. Aquacult Sci. 2007;55(2):183-189.
Crossref - Le NTT, Bach LG, Nguyen DC, et al. Evaluation of factors affecting antimicrobial activity of bacteriocin from Lactobacillus plantarum microencapsulated in alginate-gelatin capsules and its application on pork meat as a bio-preservative. Int J Environ Res Public Health. 2019;16(6):1017.
Crossref - Collado MC, Meriluoto J, Salminen S. Role of commercial probiotic strains against human pathogen adhesion to intestinal mucus. Lett Appl Microbiol. 2007;45(4):454-460.
Crossref - Lee JE, Lee NK, Paik HD. Antimicrobial and anti-biofilm effects of probiotic Lactobacillus plantarum KU200656 isolated from kimchi. Food Sci Biotechnol. 2021;30(1):97-106.
Crossref - De ADPDA, Estirpes P. Evaluation of adhesive properties of presumptive probiotic Lactobacillus plantarum strains. 2013;https://www.academia.edu/download/80481669/evaluation-of-adhesive-properties-of-presumptive-probiotic-lact_ptudZAm.pdf
- Masuda T, Hidaka A, Kondo N, Ura T, Itoh T. Intracellular enzyme activities and autolytic properties of Lactobacillus acidophilus and Lactobacillus gasseri. Food Sci Technol Res. 2005;11(3):328-331.
Crossref
© The Author(s) 2024. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.