ISSN: 0973-7510
E-ISSN: 2581-690X
The exploration of novel sources of commercially important enzymes is important due to the enormous industrial requirement. Modern isolation, screening and characterization techniques expedited such efforts. Protease is a highly specific and one of the most important primary enzymes having extensive industrial applications. It is being used in almost all the industrial sectors for one or the other hydrolyzing requirement.The aim of the present study was to isolate, screen, and performing molecular characterization of protease producing novel bacterial strain, sampled from the environmental waste soil around the Warangal region. A total of 25 samples were collected and serial dilution was done with 0.1ml of the sample, further,spreading was done on nutrient agar medium and the sample was incubated at 37⁰C for 24 to 48 hrs.Proteolytic activity in the colony-forming microorganisms was detected through skimmed milk agar medium. The results suggest that samples collected from the tannery waste soil showed high protease activity. A total of 10 colonies of TWSS-P-2 showed high protease activity through larger hydrolysis zone creation (33.5mm).Morphological and biochemical characters were studied based on their isolate and molecular characterization confirmed the strains as Rummelli Bacillus Stabekisii (TWSS-P-2).The maximum activityof the selected strains was observed at pH 7.2 and at temperature 37⁰C.We have conducted several production enhancement and optimization experiments using different carbon and nitrogen sources. Among different carbon and nitrogen sources studied, the highest protease activity of 596(U/ml) was found optimal. Our analysis confirmed that the most effective bacterial strain for the sampled soil was Rummelli Bacillus Stabekisii (WSS-P-2).
Rummelli Bacillus Stabekisii, Protease, Molecular Characterization, Environmental Sample, 16S r DNA.
Enzymes produced from microbial sources are becoming tremendously important due to their extensive and ever-growing applications in food and beverage industry1-2, pharmaceutical industry3, leather and textile industry4-5 , bioprocessing and detergent industry6, and in other relevant industries. Identification of important rate-limiting and catalytic enzymes and their large-scale production and optimization have become a routine industrial activity in the last few decades. Even though synthetic catalysts are used but the most active and efficient natural catalysts are irreplaceable and cost-effective at the same time.
Proteolytic enzymes are abundant in nature and are found in several microorganisms that can be harnessed and applied for industrial or commercial purposes. Proteases are vital for regular cellular functioning and simultaneously they have become essential in the industrial applications, occupying more than 70% of the industrial enzyme production and applications. Apart from the usual bacterial sources7 , they are also produced from fungi at an industrial scale8. Subsequently, proteases are also available from plants, animals, and viruses. This enzyme can efficiently function under a broad range of pH concentration9 which is an advantage for multiple industrial applications. The protease enzyme family is large and differs extensively in structural, functional, and biochemical properties. Such differences include target specificity, catalysismechanism, operational temperature, optimal temperature variation, pH variation, substrate specificity and stability of the enzymes. Considering their importance, they are being harvested from various sources including marine microbial organisms10. Protease enzymes are broadly classified based on their catalytic residues and substrate specificity, evolutionary conservations and variations, and optimal pH level of operation11-13. Functionally, proteases accelerate the catalysis of the peptide bonds cleaving and they are categorized into different types such as carboxypeptidases, aminopeptidases, exopeptidases, serine proteases, endopeptidases, aspartic proteases, metalloproteases, and thiol of cysteine proteases14. Functionally, a precise complex molecular system guides the specificities of each type of protease activity. Different domains and functional modules serve this purpose for the protease enzyme and provide them precise localization, substrate specificity, and required kinetic properties. Such specific non-catalytic domains are also of highly specific nature,for instance, localization is directed by archetypal sorting signals, proper enzyme activation is controlled by autoinhibitory prodomains, and contact and interaction with other proteins, substrate, receptors are managed by ancillary domains. Thus, the enzyme functions as a unit with precise functionality carried out by each domain15 Specific structural variations are also noted in different proteases along with their diversity.Bacterial proteases belong to the hydrolase category of enzymes. An earlier evolutionary analysis suggested that even after gigantic diversity,precise function of the protease family enzymes is due to the sensing activity of the nucleophile rotamer for selection of amide present in vicinity with respect to the oxyanion hole16 Thus, the stereochemistry of the catalysis is properly regulated for each protease. The enzyme displays a convergent evolution where constraint on structural evolution is predominant 16 . Industrially, it has been understood that Bacillus sp. produces the enzyme extracellularly. Analysis of the bacterial phases revealed that the production of proteases occurs in the stationary phase, specifically after the exponential phase. Media components are supposed to either induce or reduce the production of the protease enzyme.The presence of specific carbon, nitrogen, and sugar sources impact protease enzyme production.
In the present study, our aim was to screen, isolate, characterize and identify the bacterial strains that produce sample protease enzyme from the polluted soil samples. Further,the assessment was conducted to understand the optimal environment in terms of media composition, pH, and temperature to obtain an optimal yield of protease production and activity.
Following the study objective mentioned, we collected the most polluted soil samples from the surrounding of Warangal city located in the North Telangana region. The detailed methodology applied in this regard is described in the following section.
Sample sources and collection
As the preliminary requirement, all the possible sample collection sites were inspected and confirmed.Soil samples were collected from different areas of Warangal district that includes Poultry waste(referred and identified as PWSS-P) and fish waste soil samples (referred and identified as FWWS-P) from Parkal area, soil samples from dairy farm of Gudephad and Hanamkonda (referred and identified as DFSS-G), dead animal remain associated soil sample (referred and identified as DARSS-P) from Parkal municipal dump yard, soil samples from tannery wastes (referred and identified as TWSS-P-2)of Parkal region, samples from mutton wastes (referred and identified as MWSS-H) of Hanamkonda area, and The insects crushed sample (referred and identified as ICS- KU) of Kakatiya University area, an other sample from rhizosphere soil sample
(referred and identified as RSS- KU) soil of Kakatiya University Nursery. All the samples were collected from 4-5 cm depth and stored in sterile polythene bags. The sampleswere preserved properly in the site of collection and transferred into the laboratory immediately. Further, the samples were preserved at 4°C in the laboratory. Each sample containing bags were tagged with the sample collection time, date and other relevant details.
Microorganisms and culture conditions
Standard serial dilution with distilled water was performed for the samples and up to the 10-5 dilution was done.The spread plate technique was used for sample isolation.All the samples were then spread on Petri plates containing nutrient agar and incubation was done for 24 hrs. at 37°C. Morphological and biochemical characterization was done for the developed bacterial colonies after incubation to identify and confirm their species. Endospore staining was also considered to be certain on the species identity. To obtain a pure culture, the developed colonies were subcultured using nutrient agar plates. Later on, the subcultures were transferred to a different medium that was having 1% glucose, 0.5% casein, 2.5% yeast and were maintained at 37°C and at pH 7.4. Continuous shaking was applied for better mixing. All the chemicals and reagents used were purchased from High Media Company and were of analytical grade. On the basis of morphological, biochemical analysis and molecular characterization and identification, the bacterial isolates were identified17.
Screening for Protease producing bacteria
Primary screening of the organisms was done using a 1% skimmed milk agar plate where 1-gram (check the unit) of skimmed milk was added into 100 ml of Lysogeny broth (LB) medium containing agar as a solidifying agent. A total of 100 single colonies were selected and further inoculated on the skimmed milk agar plate. The incubation of the same was done for 48 hrs. at 37°C. Clear zones were identified to identify the possible casein hydrolysis18.
Protease enzyme production
Depending on the production of a clear zone in the skimmed milk agar medium, bacterial colonies were selected for protease production. Lysogeny broth (LB) medium in a 100 ml flask was used for this purpose. Continuous shaking for 48 hours was allowed at 200 rpm at a temperature of 37°C. Later on, the cultures were centrifuged at 4000 rpm at 4°C. Only the supernatant was used to assess proteolytic activity.
Protease production optimization atvarying pH concentrations and temperature
The varying pH (pH 4.0 to pH 12.0) was decided using sodium acetate (pH 5-6), sodium phosphate (pH 6- 7), simple phosphate buffer (pH 7.0) and sodium carbonate (pH 8-9). After incubation, the cell culture was treated as described above and cell-free supernatants were used for enzymatic activity19. Optimizing the production of protease is associated with pH and temperature. Therefore, to attain the optimum temperature for protease production for the considered culture, different temperatures such as 37°C, 40°C, 45°C, 55°C, and 65°C were used. The range of temperature for this study was decided to be between 20°C and 55°C for the present study. The incubation of the samples was done for 24 hrs. to 48 hrs. time period.
Detail identification of proteolytic strains
Based on the above-mentioned assessment and analysis of the colony morphology, standard Gram staining protocol and biochemical tests such as catalase, DNase and oxidase tests, the identification of bacterial strains was conducted. The identified strain was TWSS-P-2 which was further selected for additional morphological, biochemical and molecular characterization. To identify the strain properly morphologically, colony morphology and microscopic observations were done for the selected bacterial isolate TWSS-P-2. Gram staining and motility assessment were part of the analysis.
Qualitative test for protein
Assessment of the protein production was done qualitatively through the Biuret test, Ninhydrin test, Millon’s test, and Xanthoproteic test. The obtained results were determined and categorized based on the specific color production and the presence of amino acid. The total protein content was determined by Lowry’s method and the Bovine Albumin Serum was used as a control in this assessment.
Protease activity assay
Casein was dissolved in 10ml of distilled water and the pH was adjusted to 8.0 using 0.1M NaOH solution.TWSS-P-2 protease producing isolate was inoculated in LB broth and incubated at 37°C for 24 to 48 hours. Later, it was centrifuged at 8000 rpm for 15mins.The pellet was removed and the supernatant was used as a crude enzyme. The 1% casein in 0.1M phosphate buffer was considered as a substrate in this case. In this experiment, an equal volume of 1ml of enzyme and substrate was incubated at 50°C for 60mins. Later, 3ml of TCA (Tri Chloro Acetic Acid) was added and the reaction was terminated. Later, the solution was centrifuged at 5000 rpm for 15mins, then 0.5ml of supernatant was made up to 2.5ml volume adding 0.5M sodium carbonate and the solution was mixed well and incubated for 20mins at 37°C. Then 0.5ml of Folin Phenol reagent was added and the solution was inspected at 660nm using spectrophotometer. Finally, production of protease enzyme was estimated and expressed in micrograms of tyrosine released under standard assay conditions.
Molecular characterization
Molecular characterization has become an inevitable step towards the characterization of bacterial isolate and strains. The molecular marker-assisted analysis is predominant in this regard and 16S ribosomal DNA based analysis has been recognized as a standard technique. For the identification of the strains morphologically and biochemically characterized in the present study, we opted to pursue molecular characterization based on the 16S ribosomal DNA based marker. To perform the analysis, DNA was isolated from the culture initially, followed by a detail quality evaluation using 1.0% agarose gel. Fragment of 16S ribosomal DNA gene was amplified using 27F and 1492R primers. Further, the PCR amplicon was purified to remove the present contaminants. Following the standard protocol, forward and reverse DNA sequencing reaction of PCR amplicon was carried out using the respective primers.Sequencing was done through BDT v3.1 Cycle sequencing kit on ABI 3730xl Genetic Analyzer.
Sequence analysis and phylogenetic analysis
A total of 6 different sequences as part of the six frames were generated using the respective forward and reverse primers. Later consensus sequence of the sample was generated from forward and reverse sequence data using multiple alignment software program Clustal W. The 16S rDNA gene sequence was used to carry out BLAST20 analysis against the NCBI GenBank database. Obtained best 10 hits were used for further multiple sequence alignment using ClustalW21 software with default gap opening and gap extension penalty.
After identification of the sequence, and collecting the top hits with higher proximity, phylogenetic analysis was conducted using the MEGA tool (version 7.0)22. The required distance matrix was generated for the considered top 10 sequences obtained from the BLAST analysis.
All the experimentations conducted in this study led to the successful outcome of identification of the most potent bacterial strain that is having maximum protease producing capability. The isolated, characterized and identified strain could be of immense industrial importance in the future. The obtained results of the experimentation are described in detail in the following section.
The analysis outcome of this study is presented following the order of the received results using the well-established logical experimentation flow.
Microorganism culture and isolation
As mentioned in the methodology section, samples were collected from multiple sites having a varying degree of environmental pollutions and contamination in the soil. A total of 25 samples were considered for the study that was diluted through serial dilution up to the 10-5 to 10-6 level. During the study, a total of 75 Petri plates and 300 bacterial colonies were developed initially to isolate the most important bacterial strain that can have a maximum level of protease activity. All these cultures were consecutively subjected to subcultures on a regular basis to obtain pure cultures.All the isolated 300 pure bacterial cultures were screened for protease activity using skimmed milk agar medium as discussed earlier and the positive colonies were spotted. The skimmed milk agar with a volume of 1000 ml was developed using casein enzyme hydrolysate (5grams), Yeast extract (2.5grams), Dextrose sugar (1gram), skim milk agar powder (28grams), Agar (15grams), anddistilled water (1 liter). The pH was maintained at pH level of 7.2. The skimmed milk agar preparation was further sterilized through autoclave at 15 lbs. pressure at 121°C for 15 mins. Plating of the pure culture bacterial strains were done through streaking the loop on a skimmed milk agar plate. All the inoculated plates were incubated at 37°C for 24 hrs.Followed by incubation, visual inspection suggested that 66 isolates out of 300 pure culture showed excellent protease activity through acceptable zone of hydrolysis. Out of the 66 isolates that showed clear zone of hydrolysis, the bacterial strains from the sample of poultry waste (PWSS-P) were found to have zone of hydrolysis of 3 mm, 6mm,10mm, 12mm,15mm,21mm,23mm,25mm, 27mm,30 mm, and 31mmdiameter respectively. Similarly, the promising strains from the fish waste soil samples (FWWS-P) displayed hydrolysis zone of 9mm, 11 mm, 13mm, 16mm, 19mm, 23mm, 26mm, 29mm, 31mm, 35mm and 42mm respectively. The analysis outcome for the samples of dairy farm wastes (DFSS-H) showed hydrolysis zone of 1mm, 5mm, 9mm, 12mm, 15mm, 19mm, 22mm, 25mm, 28mm, 33mm and 36mmrespectively. Strains present in the samples of dead animals remain associated (DARSS-P) soil sample showed the hydrolysis zones of 2 mm, 7mm, 11mm, 14mm, 17mm, 21mm, and 28mm respectively. For the samples collected from tannery wastes (TWSS-P), the strains displayed the hydrolysis zones of 2 mm,7 mm, 8 mm, 11mm, 15mm,16mm,21mm, and 30mm respectively. Obtained bacterial strains from the mutton wastes (MWSS-H), provided hydrolysis zone of 1 mm, 3 mm, 7 mm,10 mm,16 mm,17 mm, and 19 mm. Likewise, the strains isolated from the samples from the crust insects polluted (ICS-KU and RSS-KU) soil showed zone of hydrolysis of 2 mm,5mm,6mm,8mm,10mm,12mm and 4mm,8mm,9mm,10mm respectively.Thorough inspection all outcomes and comparison suggested that maximum zone of hydrolysis was provided by the isolate TWSS-P-2 with 33.5 mm whereas the lowest result was obtained from RSS-KU-9 with 2 mm diameter.The difference (D-d) between the diameter of the total clear hydrolysis zone including colony (D) and the diameter of the colony (d) was taken as a measure to calculate the specific protease activity. The positive isolate that showed maximum zone of hydrolysis (TWSS-P-2) was chosen for further studies and maintained on nutrient agar slants at 4°C.
Morphological and microscopic examination
Morphological analysis was performed for the selected TWSS-P-2 isolate. The obtained qualitative result of the morphological characterization of the selected bacterial isolate PWS-6 is provided in Table 1. Assessment of the colony morphology suggested that the size of the isolate was found to be medium and big, and the shapes were irregularly forming a rough surface. The pigmentation analysis resulted in white, yellow and pink. Other morphological characters are given in Table 1.
Table (1):
Morphological characteristics of isolate TWSS-P-2 on a nutrient agar plate.
Characteristic Colony Morphology |
Results |
|---|---|
Size |
Medium |
Shape |
Regular |
Surface |
Rough |
Elevation |
Flat |
Texture |
Moist |
Pigmentation |
White |
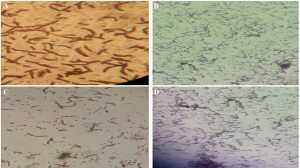
A detail microscopic analysis suggested that all bacteria of TWSS-P-2 were gram-positive in nature and endospore staining was also found positive along with the motile nature of the bacteria in the colony. The obtained morphological views of the bacteria are represented in Fig. 1. Morphological observations of the developed colonies after incubation allowed identifying the samples as Bacillus species.
Biochemical characterization outcome
Biochemical characterization is of great significance to understand the characteristics of bacterial species. Altogether, 14 different biochemical assessments were done for the samples. The results obtained for biochemical characteristics analysis of the selected bacterial isolate TWSS-P-2 are presented in Table 2. The analyses suggested that the bacteria present in this sample were found to be catalase test, nitrate reduction test, Urease production test, VoguesProskauer test, Hydrogen sulfide test, Nitrogen production test, Carbohydrate fermentation test, and Starch utilization test positive whereas negative outcome was found for Oxidase test, Methyl red test, Indole test, H2s production test, and Citrate test.
Table (2):
Biochemical characterization of TWSS-P-2.
S.NO |
Biochemical test |
Result |
|---|---|---|
1 |
Catalase test |
+ve |
2 |
Oxidase test |
-ve |
3 |
Indole test |
-ve |
4 |
Methyl red |
-ve |
5 |
Nitrate reduction test |
+ve |
6 |
H2s production test |
-ve |
7 |
Urease production test |
+ve |
8 |
Voges Proskauer test |
+ve |
10 |
Hydrogen sulfide test |
+ve |
11 |
Nitrogen production test |
+ve |
12 |
Carbohydrate fermentation test |
+ve |
13 |
Citrate test |
-ve |
14 |
Starch utilization test |
+ve |
The representation of some of the biochemical test is provided in Fig. 2.
Fig. 2. (A) Qualitative test for Std.Protein and Test Sample.(B) Quntitative Test for Protein by Lowry´s Method.
Protease production and activity estimation
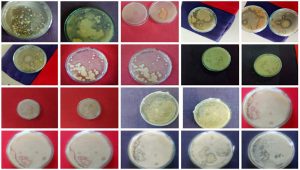
As described in the methodology section, the estimation of the protease production was done and the result obtained for protein production assay is presented in Fig. 2. As mentioned earlier the protease production colonies were selected for further analysis, a representation of such colonies is provided in Fig. 3. The selected bacterial isolate TWSS-P-2 showed high protease activity, based on the morphological and biochemical characteristics. Indication of casein hydrolysis was decided through the obtained clear zones18 (Olajuyigbe and Ajele, 2005).
The observed results of the protease activity assay are provided in Fig. 4.
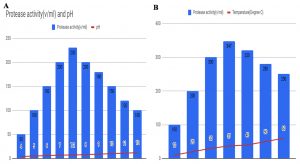
Protease activity was optimized for the selected strains with varying pH and temperature with the goal of optimum production. Fig. 5 shows the output of protease activity in different pH and temperature. The experiment for pH variation was conducted from pH 4.0 to 12.0 levels (Fig. 5A) and for temperature, the values were considered from 10°C to 60°C (Fig.5B). It was noted that in case of pH variation, maximum output of 230 U/ml protease activity was obtained at pH 7.2 which was nearly neutral and for the temperature parameter to obtain the best results of 347 U/ml were observed at 37°C. Hence, the results suggest the almost neutral pH and 37°C may yield the best outcome for protease activity of the selected bacterial species.
Fig. 5. Optimization of protease activity with respect to (A) varying pH, and (B) varying temperature for the selected bacterial strain.
In addition to the optimization analysis, we also tested the efficiency of the protease activity of the TWSS-P-2 strain in the presence of different carbon sources as presented in Table 3. It was interesting to witness the best results of 596 U/ml of protease activity in the presence of glucose which was followed by maltose (509 U/ml). The presence of glycerol and starch showed very less sensitivity towards protease activity as represented in Table 3.
Table (3):
Estimated protease activity of the selected TWSS-P-2 strain in the presence of different carbon sources.
| Bacterial strain | Carbon sources | Protease activity(U/mL) |
|---|---|---|
| TWSS-P-2 | Lactose | 464 |
| galactose | 391 | |
| Maltose | 509 | |
| Glucose | 596(high) | |
| Glycerol | 153 | |
| starch | 186 |
The stimulation of different nitrogen sources was also explored as presented in Table 4. It was observed that presence of casein was having good impact on the protease activity for TWSS-P-2 strains (39.09 U/ml) whereas least effect was found in the presence of yeast extract and peptone as shown in Table 4.
Table (4):
Estimated protease activity of the selected TWSS-P-2 strain in the presence of different nitrogen sources.
S.No. |
Nitrogen sources |
Protease activity(U/mL) |
|---|---|---|
TWSS-P-2 |
(NH₄)₂SO₄ Casein Yeast extract Beef extract Peptone |
19.35 39.09(high) 3.09 20.05 9.58 |
Molecular characterization
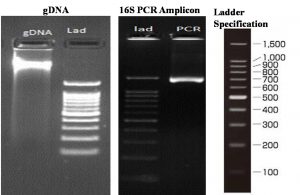
Ribosomal marker DNA assisted analysis was performed to attain the molecular characterization of the selected sample as described in the methodology section. 16S rDNA gene amplification was opted to fulfill the objective of this study. This specific marker is extensively used in bacterial species identification and understanding the evolutionary position of various bacterial strains. Several novel strains have been discovered using this 16S rDNA region of the genomes. Fig. 6 represents the analysis details where genomic DNA against the 100bp ladder (Promega, USA), 16S PCR amplicon and ladder, and detail ladder specification is represented. A single band of high-molecular-weight DNA has been observed which was corresponding to the discrete PCR amplicon band of 1500 bp.
Fig. 6. Molecular characterization of the bacterial strains through 16S rDNA. (A) View of the genomic DNA and the Ladder, (B) 16S PCR Amplicon, and (C) The opted ladder specifications.
Sequence analysis and Phylogenetic analysis results
The obtained output of the agarose gel electrophoresis was used for sequencing using the ABI 3730xl Genetic Analyzer. Following the protocol, 6 frame translations were carried out and 3 forward and 3 reverse frame sequences were generated. The aligner tool was used to obtain the consensus sequence from the generated 6 sequences. The consensus sequence was of 1405 nucleotide length with the abundance of “G” and “A” nucleotides of 30.46% and 25.12% respectively followed by 21.35% “T” and 23.06% “C” nucleotides in the sequence. The estimated molecular weight of the sequence was ~17.01 Da. Nucleotide-nucleotide BLAST analysis of the sequenceopted and the result of the top 10 sequences was identified and collected based on the total score and expected value (E-value). High similarity was observed with Rummelii Bacillus Stabekisiias represented in Table 5. The top 2 hits showed 100% query coverage and 0 E-value with more than 99.5% percent identity (Table 5) confirming Rummelii Bacillus Stabekisii as the designate species for the considered sequence.
Table (5):
Sequences producing significantalignments in BLAST analysis.
Description |
Max Score |
Total Score |
Query Coverage |
E value |
Per. Ident |
Accession |
|---|---|---|---|---|---|---|
Rummelii bacillus stabekisii strain NBRC 104870 16S ribosomal RNA, partial sequence |
2582 |
2582 |
100% |
0 |
99.86% |
NR_114270.1 |
Rummelii bacillus stabekisii strain KSC-SF6g 16S ribosomal RNA, partial sequence |
2579 |
2579 |
100% |
0 |
99.79% |
NR_043992.1 |
Rummelii bacillu spycnus strain NBRC 101231 16S ribosomal RNA, partial sequence |
2429 |
2429 |
100% |
0 |
97.87% |
NR_041521.1 |
Rummelii bacillus suwonensis strain G2016 Sribosomal RNA, partial sequence |
2320 |
2320 |
99% |
0 |
96.64% |
NR_109749.1 |
Kurthiagibsonii strain NBRC 15534 16S ribosomal RNA, partial sequence |
2320 |
2320 |
99% |
0 |
96.71% |
NR_041520.1 |
Kurthiagibsonii strain NCIMB 9758 16S ribosomal RNA, partial sequence |
2302 |
2302 |
99% |
0 |
96.49% |
NR_119002.1 |
Kurthiasibirica strain NBRC10153016 Sribosomal RNA, partial sequence |
2300 |
2300 |
99% |
0 |
96.42% |
NR_112626.1 |
Kurthiasibirica strain 13-2216 Sribosomal RNA, partial sequence |
2300 |
2300 |
99% |
0 |
96.42% |
NR_042214.1 |
Viridi bacillus arvi strain LMG2216516 Sribosomal RNA, partial sequence |
2300 |
2300 |
100% |
0 |
96.17% |
NR_025627.1 |
Kurthia populi strain 10y-1416 Sribosoma lRNA, partial sequence |
2298 |
2298 |
99% |
0 |
96.43% |
NR_144587.1 |
Phylogenetic Tree construction
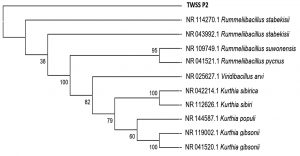
To obtain the phylogenetic relationship of the top 10 sequences ClustalW was used to develop the multiple sequence alignment using the default gap opening penalty and gap extension penalty values for nucleotides. The evolutionary analysis was done using the Maximum Likelihood method and the model selected was Kimura 2-parameters. To obtain better consensus 1000 bootstrap replicates were used. All the branches that were unable to reproduce more than 50% replicates in bootstrap analysis were collapsed. The replicate percentages are represented along with the branches in an optimized scale of 0 to 100(Fig. 7). High bootstrap replicates are evident from the tree. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach and then selecting thetopology with superiorlog likelihood value. The analysis involved 11 nucleotide sequences. Codon positions included were 1st+2nd+3rd+Noncoding. All positions containing gaps and missing data were eliminated. There were a total of 1434 positions in the final dataset. Evolutionary analyses were conducted in MEGA7. Such advanced computational techniques have been regularly used to understand the commercially important enzymes.
Fig. 7. Phylogenetic tree reconstructed applying maximum likelihood method and 1000 bootstrap replications.
The evolutionary distances obtained for the considered sequences are presented in Table 6. The study provided an excellent outcome where we were successful in isolating and screening the important bacterial strains from various waste and polluted soil samples. Out of all, we found that the intended bacterial species with high protease activity was found in the tannery waste and dead animal wastes as mentioned in the result section. Moreover, optimization of the same for protease activity released that near-neutral pH and 37°C temperature was preferred by the presently isolated strains for optimal functionality.
Table (6):
Estimates of evolutionary divergence between the sequences considered for this study.
TWSS_P_2 |
0.000 |
0.000 |
0.003 |
0.005 |
0.005 |
0.005 |
0.005 |
0.005 |
0.005 |
0.005 |
|
NR_114270.1 |
0.000 |
0.000 |
0.003 |
0.005 |
0.005 |
0.005 |
0.005 |
0.005 |
0.005 |
0.005 |
|
NR_043992.1 |
0.000 |
0.000 |
0.003 |
0.005 |
0.005 |
0.005 |
0.005 |
0.005 |
0.005 |
0.005 |
|
NR_041521.1 |
0.019 |
0.019 |
0.019 |
0.004 |
0.005 |
0.005 |
0.006 |
0.006 |
0.004 |
0.005 |
|
NR_109749.1 |
0.035 |
0.035 |
0.035 |
0.019 |
0.006 |
0.005 |
0.006 |
0.006 |
0.005 |
0.006 |
|
NR_041520.1 |
0.034 |
0.034 |
0.034 |
0.038 |
0.046 |
0.001 |
0.004 |
0.004 |
0.005 |
0.005 |
|
NR_119002.1 |
0.035 |
0.035 |
0.035 |
0.039 |
0.047 |
0.001 |
0.004 |
0.004 |
0.004 |
0.005 |
|
NR_112626.1 |
0.035 |
0.035 |
0.035 |
0.039 |
0.049 |
0.026 |
0.027 |
0.000 |
0.005 |
0.005 |
|
NR_042214.1 |
0.035 |
0.035 |
0.035 |
0.039 |
0.049 |
0.026 |
0.027 |
0.000 |
0.005 |
0.005 |
|
NR_025627.1 |
0.035 |
0.035 |
0.035 |
0.030 |
0.039 |
0.034 |
0.035 |
0.035 |
0.035 |
0.005 |
|
NR_144587.1 |
0.039 |
0.039 |
0.039 |
0.037 |
0.045 |
0.031 |
0.032 |
0.035 |
0.035 |
0.041 |
The bacterial species and strain identified to be the most potent protease producer is Rummelii Bacillus Stabekisii. Recently, the whole genome of this bacterial species has been sequenced for the PP9 isolated from the Antarctic soil23. This particular bacterium has shown to survive in extreme or harsh environmental conditions, therefore, the stability of the enzyme isolated from this species could be more. Earlier phylogenetic studies suggested that this bacterial species is close to Bacillus pycnus24.
Identification of novel microbial sources to meet the industry requirement is important and an ongoing effort. Our experiments allowed us to identify a Rumellii Bacillus Stabikisii with considerable production capabilities of protease enzyme. The whole-genome of this strain has been sequenced recently done24 and our exploration of the high protease producing feature of this strain may increase attention towards this particular bacterial strain. Moreover, additional gene and protein-based analysis can be attempted to understand such productivity of enzymes at the molecular level. According to our understanding this could be a pioneer report on this bacterial species as an excellent protease enzyme producer.
Identification of novel bacterial species is of great importance, especially for the purpose of industrial enzyme production. Exploration of novel sources is required for better, highly active, specific, and cost-effective enzyme production. Environmental wastes could serve as an excellent cheap resource for this purpose. It can provide us with stress tolerable bacterial strains and enzymes and also help in treating and managing the ever-growing wastes. We report a unique source of protease enzyme from the waste and polluted soil samples of Warangal region. Our study presents successful isolation and optimization of protease enzyme from Rummelii Bacillus Stabekisii, a highly tolerant bacterial strain. Upscaling of this strain may yield better industrial outcomes for the protease production from microbial sources in the future.
ACKNOWLEDGMENTS
We would like to express our heartful thanks to Head Department of Biotechnology for providing laboratory facilities. Thanks to Eurofins Genomics India Pvt Lt. for providing assistance on bioinformatics analysis. The authors are grateful to Dr. P. Srinivas for English editing of the manuscript.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
This work was supported by the University Grants Commission. (Grant number: no.f. / 2015-17/RGNF-2015-17-TEL-1072).
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
AVAILABILITY OF DATA
The datasets generated and/or analyzed during the current study are available in the NCBI database repository, Accession No: MN- 163058
- Marquez OH, Serrano MF, Bonilla MP, Sandoval MB, Alameda EJ, Gonzalez GL. Activity and Stability in the Presence of a non-Ionic Surfactant of a Protease for Hard Surface Cleaning in Food Industry. Chemical Engineering Transactions, 2019; 15: 75:187-92.
- Zhang Y, He S, Simpson BK. Enzymes in food bioprocessing—novel food enzymes, applications, and related techniques. Current Opinion in Food Science, 2018; 19: 30-5.
Crossref - McKerrow JH. Update on drug development targeting parasite cysteine proteases. PLoS Neglected Tropical Diseases. 2018; 12(8): e0005850.
Crossref - Zhou C, Qin H, Chen X, Zhang Y, Xue Y, Ma Y. A novel alkaline protease from alkaliphilic Idiomarina sp. C9-1 with potential application for eco-friendly enzymatic dehairing in the leather industry. Scientific Reports, 2018; 8(1): 16467.
Crossref - Benmrad MO, Moujehed E, Elhoul MB, Mechri S, Bejar S, Zouari R, Baffoun A, Jaouadi B. Production, purification, and biochemical characterization of serine alkaline protease from Penicillium chrysogenium strain X5 used as excellent bio-additive for textile processing. International Journal of Biological Macromolecules, 2018; 119: 1002-16.
Crossref - Schroder M, Lenting HB, Kandelbauer A, Silva CJ, Cavaco-Paulo A, Gubitz GM. Restricting detergent protease action to surface of protein fibres by chemical modification. Applied Microbiology and Biotechnology, 2006; 72(4): 738-44.
Crossref - Contesini FJ, Melo RR, Sato HH. An overview of Bacillus proteases: from production to application. Critical Reviews in Biotechnology, 2018; 38(3): 321-34.
Crossref - Mandal S, Banerjee D. Proteases from Endophytic Fungi with Potential Industrial Applications. InRecent Advancement in White Biotechnology Through Fungi, 2019 (pp. 319-359). Springer, Cham.
Crossref - Sawant R, Nagendran S. Protease: an enzyme with multiple industrial applications. World Journal of Pharmacy and Pharmaceutical Sciences, 2014; 3(6): 568-79.
- Barzkar N, Homaei A, Hemmati R, Patel S. Thermostable marine microbial proteases for industrial applications: scopes and risks. Extremophiles, 2018; 22(3): 335-46.
Crossref - Garcia-Carreon FL. Classification of proteases without tears. Biochemical Education, 1997; 25(3): 161-7.
Crossref - Barrett AJ. Proteases. Current protocols in protein, Science, 2000; 21(1): 21-1.
Crossref - Nishimura K, Kawamura Y, Matoba T, Yonezawa D. Classification of proteases in Antarctic krill. Agricultural and Biological Chemistry, 1983; 47(11): 2577-83.
Crossref - Rani K, Rana R, Datt S. Review on latest overview of proteases. Int J Curr Life Sci., 2012; 2(1): 12-8.
- Lopez-Otin C, Bond JS. Proteases: multifunctional enzymes in life and disease. Journal of Biological Chemistry, 2008; 283(45): 30433-7.
Crossref - Buller AR, Townsend CA. Intrinsic evolutionary constraints on protease structure, enzyme acylation, and the identity of the catalytic triad. Proceedings of the National Academy of Sciences, 2013; 110(8): E653-61.
Crossref - Sneath PH, Mair NS, Sharpe ME, Holt JG. Bergey’s manual of systematic bacteriology. Williams & Wilkins, 1986; 2.
- Olajuyigbe FM, Ajele JO. Production dynamics of extracellular protease from Bacillus species. African Journal of Biotechnology, 2005; 4(8): 776-9.
- Sevinc N, Demirkan E. Production of protease by Bacillus sp. N-40 isolated from soil and its enzymatic properties. J Biol Environ Sci., 2011; 5(14): 95-103.
- Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL. NCBI BLAST: a better web interface. Nucleic Acids Research, 2008; 36(suppl_2): W5-9.
Crossref - Thompson JD, Gibson TJ, Higgins DG. Multiple sequence alignment using ClustalW and ClustalX. Current Protocols in Bioinformatics, 2003; (1): 2-3.
Crossref - Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 2016; 33(7): 1870-4.
Crossref - da Mota FF, Vollu RE, Jurelevicius D, Seldin L. Whole-genome sequence of Rummeliibacillusstabekisii strain PP9 isolated from Antarctic soil. Genome Announc, 2016; 4(3): e00416-16.
Crossref - Vaishampayan P, Miyashita M, Ohnishi A, Satomi M, Rooney A, La Duc MT, Venkateswaran K. Description of Rummeliibacillusstabekisii gen. nov., sp. nov. and reclassification of Bacillus pycnus Nakamura et al. 2002 as Rummeliibacilluspycnus comb. nov. International Journal of Systematic and Evolutionary Microbiology. 2009; 59(5): 1094-9.
Crossref
© The Author(s) 2020. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.