ISSN: 0973-7510
E-ISSN: 2581-690X
The amount of petroleum hydrocarbons affects biodegradation. Different classifications can be used to petroleum components Asphaltenes, ethers, fatty acids, porphyrins and resins saturated fatty acids. Twenty soil samples tainted with petroleum products and crude oil were gathered from the Basra oil fields and other parts of the Babylon governorate; of these twenty samples, seventeen showed Pseudomonas aeruginosa (P. aeruginosa) growth on the chromogenic agars, which manifested as purple colonies. Using 16S rRNA and PCR technology with a 505 bp PCR product size, P. aeruginosa isolated from soil contaminated with petroleum products was identified. The Kirby-Bauer disk diffusion susceptibility test, a method for testing antibiotic sensitivity, produced the following findings. The percentages of ciprofloxacin CIP, imipenem IPM, meropenem MEM, tobramycin TOB and amoxicillin + clavulanic acid AMC equal to 10 mg, ceftazidime CAZ (30 mg), and cefotaxime CTX (30 mg) are 59%, 70%, and 82% respectively. The goal of creating a minimum salt media and introducing diesel at 0.05% concentration is to ascertain how well bacteria may grow in petroleum product-containing settings by breaking down diesel. For a total of 21 days, the optical density in this test is measured every seven days. The results show a rise in the optical density, which suggests that the culture medium being used is growing more quickly. Additionally, it was noted that the center’s color and turbidity was altered. P. aeruginosa can grow in soil contaminated with petroleum products, had the ability to degrade petroleum pollutant, highly resisted to antibiotic that used in this study.
Pseudomonas aeruginosa, PCR, Antibiotic Sensitivity, Soil Bacteria, Petroleum, Biodegradation
Biodegradation is influenced by petroleum hydrocarbon concentration.1,2 Different classifications can be used for petroleum components. Asphaltenes, ethers, fatty acids, porphyrins, and resins3,4 saturated fatty acids. The following hydrocarbons have less effectiveness with microbial degradation when it comes to reducing structural complexity: the alkane-like compounds in a tree, Small-scale aromatic compounds, and Fragrant polymers. Pseudomonas, Nocardia, Xanthomonas, Bacterium, Corynebacterium, Mycobacterium, and Acinetobacter are among the rare bacteria that can completely oxidize aliphatic hydrocarbons.5,6 Benzene rings and the structural complexity of aromatic hydrocarbons are factors that affect biodegradability.7
High molecular weight polycyclic aromatics can be inefficiently metabolized by certain bacteria.8-10 Because microorganisms oxidize petroleum components preferentially, cleaning requires the presence of bacteria and fungi. Soil tainted with petroleum provides microbiological cultures. Four isolates from an oil mine in northern Ecuador, two fungal (Geomyces sp. strain HV) and two bacterial (Bacillus thuringiensis B3 and B. cereus B6), may be useful for bioremediation of soil contaminated by crude oil.11 Biodegradation is influenced by microbial species, pH, temperature, aeration, water availability, and biogenic minerals. Petroleum breakdown by bacteria is influenced by temperature. The optimal temperature range for petroleum biodegradation in the soil is between 30 and 40 °C. This is hotter than the summertime 25-30 °C in soil from middle latitudes.12,13 High temperatures kill a lot of bacteria by destroying their biological structures. Cellular metabolism is slowed by low temperatures. Petroleum biodegradation is slowed down if the temperature is not right14 Enzymatic oxidation mediated by oxygenase is the first step in the breakdown of hydrocarbons.15,16 Anaerobic environments, however, may also result in deterioration.17 Electrons can be received by ferric ions and nitrate. Fuel oil18 toluene19 and even low temperatures20,21 bioremediated petroleum-contaminated soils.22
The processes involved in microbial bioremediation are intricate and multifaceted. Numerous advantages come from the wide variety of microorganisms and their various hydrocarbon detoxification techniques. Numerous pollutants in the soil are eliminated by bioremediation. To protect the ecosystem, petroleum and its byproducts must be removed from contaminated soil. According to this summary, bioremediation of petroleum-contaminated soil is effective and has a lot of promise. Alkanes can be broken down by many bacteria; however, the process is highly dependent on branch topologies and the length of the carbon chain. More research is required on alkene and aromatic degradation. Biosurfactants are of practical interest in bioremediation. Plants and bacteria together have potential. Due to the complexity of microbial bioremediation and the ways in which environmental factors and pollutant types influence their effectiveness, many study findings are not used. We must get past this obstacle. The study and uses of petroleum bioremediation are the main emphasis of applied ecology, or environmental science. This study aims to isolate Pseudomonas aeruginosa present in soils contaminated with petroleum products, and to evaluate the efficiency of these isolates in eliminating contamination.
Sample collection
Using sterile instruments, twenty soil samples were taken at a depth of roughly 10 cm. Approximately 100 cc of soil was taken and stored in sterilized containers. Samples of soil were collected from several areas inside the Hilla Governorate as well as from the Basra crude oil fields, as shown in Table 1.
Table (1):
MSM content
No. |
Per liter |
Weight |
|---|---|---|
1. |
FeSO4 |
1 mg |
2. |
MgSO4.7H2O |
200 mg |
3. |
Na2HPO4 |
210 mg |
4. |
NaH2PO4 |
90 mg |
5. |
CuSO4.5H2O |
5 µg |
6. |
H3Bo3 |
10 µg |
7. |
MnSO4.5H2O |
10 µg |
8. |
ZnSO4.7H2O |
70 µg |
9. |
MoO3 |
10 µg |
10. |
CoSO4 |
10 µg |
11. |
KCl |
40 mg |
12. |
CaCl2 |
15 mg |
13. |
NH4Cl |
500 mg |
14. |
NaNO3 |
2 mg |
15. |
0.05% (v/v) diesel |
Culture cultivation
One gram of soil was collected from each sample, mixed with nine milliliters of brain heart infusion media, and incubated for twenty-seven hours at thirty-seven degrees Celsius. Next, a loop complete transfer was obtained from each sample, plotted on the chromogenic agar medium for P. aeuroginosa, and cultured for twenty-seven hours at thirty-seven degrees Celsius.
Antibiotic susceptibility
We completed the disk diffusion of susceptibility tests in accordance with (CLSI 2023) requirements. Ciprofloxacin CIP (10 mg), Imipenem IPM (10 mg), Meropenem MEM (10 mg), Tobramycin (10 mg), Amoxicillin + clavulanic acid (10 mg), Ceftazidime CAZ (30 mg), and Cefotaxime CTX (30 mg) were the antibiotics disc potency that were administered. Every test was conducted on the Muller-Hinton Agar platform.
Identifying the activity of microorganisms in diesel
Inoculate colonies of different bacteria (previous experiment) into 50 ml mineral salts medium (MSM).
The growth response of each of the above isolated bacteria on diesel can initially determine at 7 days intervals by physical appearance (turbidity) and measuring the optical density (O.D.) at 540 nm, after an incubation period of 12 days at 28 °C.
Molecular identification of Candida species
The PCR test was performed using the primer in Table 2 and in accordance with the guidelines listed in Table 3 to identify the isolated species from the petroleum product-contaminated soil.
Table (2):
Primer sequence 5′-3′
| Gene | Direction | Sequence 5′-3′ | Product size |
|---|---|---|---|
| 16S rRNA | F | TGCCTGGTAGTGGGGGATAA | 505 bp |
| r | GGATGCAGTTCCCAGGTTGA |
Table (3):
PCR condition for amplifying C and gene
| No. | Steps | Temp. (°C) | Time | Cycles |
|---|---|---|---|---|
| 1. | Initial Denaturation | 95 | 3 min. | 35 cycle |
| 2. | Denaturation | 95 | 30 sec. | |
| 3. | Annealing | 66 | 30 sec. | |
| 4. | Extension | 72 | 45 sec. | |
| 5. | Final Extension | 72 | 5 min. | |
| 6. | Storage | 4 |
The mixture of PCR was 2 µl of DNA sample, 1.5 µl of each forward and reverse primer, and 15 µl of green master mix (Promega).
Isolation
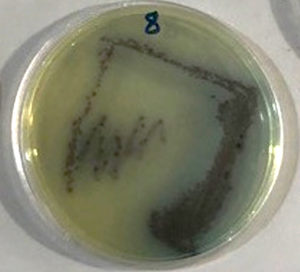
Twenty soil samples tainted with petroleum products and crude oil were gathered from the Basra oil fields and various parts of the Babylon governorate; of these twenty samples, seventeen showed P. aeruginosa growth on the chromogenic agars, manifesting as purple colonies (Figure 1).
Molecular identification of Candida species
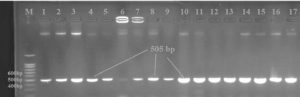
Using PCR technology and a 16S rRNA Table 1, P. aeruginosa isolated from soil contaminated with petroleum products was identified in this study. The PCR product size was 505 bp (Figure 2). It was possible to isolate a highly efficient hydrocarbon-degrading strain from a lake wetland. When peptone and beef extract are present, this strain develops swiftly. The colonies in Figure 1 have boundaries that expand unevenly and are colored yellow brown. In addition, this strain gives the MSM solution a subtle pink hue. According to SEM analysis, this strain has a length and width of around 0.4 and 1.0 meters, and it is Gram-negative. The 16S rRNA gene sequence has 1389 base pairs and a G+C content of 54.2%. About 100% of the 16S rRNA gene segment resembles Pseudomonas aeruginosa.
Figure 2. Agarose gel electrophoresis of PCR for 16S rRNA gene; M: Marker DNA ladder; lane 1-17 samples; 1.5% agarose gel with safe red stain, at 80 V/cm, it took sixty minutes
Antibiotic susceptibility
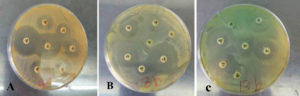
The isolates’ resistance rates were found to be extremely high when an antibiotic sensitivity test utilizing disc (Kirby-Bauer disk diffusion susceptibility test) was performed, as indicated in Table 4 and Figure 3.
Figure 3. Antibiotic susceptibility of P. aeruginosa. (A) P. aeruginosa isolate No. 2 which was sensitive to all antibiotics used, (B) P. aeruginosa isolate No. 8 which was resistant to three types of antibiotics, (C) P. aeruginosa isolate No. 13 resistant to two types of antibiotics that used
Table (4):
Antibiotic susceptibility of P. aeruginosa
No. |
Antibiotic |
Resistance % according to CLSI 2023 |
|---|---|---|
1. |
Ciprofloxacin CIP (10 mg) |
59 |
2. |
Imipenem IPM (10 mg) |
70 |
3. |
Meropenem MEM (10 mg) |
70 |
4. |
Tobramycin TOB (10 mg) |
82 |
5. |
Amoxicillin + clavulanic acid AMC (10 mg) |
35 |
6. |
Ceftazidime CAZ (30 mg) |
35 |
7. |
Cefotaxime CTX (30 mg) |
35 |
Identifying the activity of microorganisms in diesel
The purpose of making a minimum salt media and adding 0.05% diesel to it is to test how well bacteria can grow in petroleum product-containing settings by breaking down desal. For a duration of 21 days, the optical density in this test is evaluated every seven days. The results show a rise in the optical density, which suggests that the culture medium being used is growing more quickly (Table 5). Additionally, Figure 4 showed that the center’s color and turbidity had changed.
Table (5):
The change in optical density over three weeks for different isolates
Candida species |
After 7 days |
After 14 days |
After 21 days |
|---|---|---|---|
0.05% msm with desal |
1.694 |
– |
– |
Isolate 1 |
0.967 |
1.394 |
1.894 |
Isolate 2 |
0.62 |
1.439 |
1.939 |
Isolate 3 |
0.227 |
1.294 |
1.794 |
Isolate 4 |
0.815 |
1.305 |
1.805 |
Isolate 5 |
0.675 |
1.431 |
1.931 |
Isolate 6 |
0.395 |
1.166 |
1.666 |
Isolate 7 |
0.284 |
1.194 |
1.694 |
Isolate 8 |
0.347 |
0.352 |
1.349 |
Isolate 9 |
0.103 |
1.388 |
0.852 |
Isolate 10 |
0.091 |
1.401 |
1.888 |
Isolate 11 |
0.865 |
1.302 |
1.901 |
Isolate 12 |
0.967 |
1.394 |
1.802 |
Isolate 13 |
0.613 |
1.439 |
1.894 |
Isolate 14 |
0.967 |
1.294 |
1.939 |
Isolate 15 |
0.62 |
1.305 |
1.794 |
Isolate 16 |
0.227 |
1.431 |
1.805 |
isolate 17 |
0.815 |
1.166 |
1.931 |
Water and soil samples from several hydrocarbon-contaminated sites in Basrah were found to contain Bacillus spp., Pseudomonas spp., and Micrococcus spp.23 P. aeruginosa was one of the nine bacterial strains that Kridi and her colleagues were able to recover. Based on biochemical testing and unique morphological features, P. aeruginosa was identified.24
In a molecular study, it was possible to isolate a highly efficient hydrocarbon-degrading strain from a lake wetland. When peptone and beef extract are present, this strain develops swiftly. The colonies in Figure 1 have boundaries that expand unevenly and are colored yellow brown. In addition, this strain gives the MSM solution a subtle pink hue. According to SEM analysis, this strain has a length and width of around 0.4 and 1.0 meters, and it is Gram-negative. The 16S rRNA gene sequence has 1389 base pairs and a G+C content of 54.2%. About 100% of the 16S rRNA gene segment resembles Pseudomonas aeruginosa, indicating a stable genetic clade with previously identified Pseudomonas strains.25
Antibiotic sensitivity, Sedighi’s research showed that P. aeruginosa populations that produce MBL are a major hazard to therapeutics. The rate at which MBL is causing imipenem resistance has skyrocketed. According to Kadivarian et al.26 early detection and infection control measures are the most effective antimicrobial methods for this bacterium.26
Baban’s work aims to limit the overuse of carbapenem antibiotics by developing antimicrobial stewardship. Early identification of isolates resistant to carbapenem is essential to reducing the possibility of these isolates spreading to patients in critical condition. Active surveillance and rigorous adherence to infection prevention and control measures may be useful tactics for reducing the emergence of carbapenemase resistance because of Baban’s work.27
Liu and colleagues obtained a novel strain of P. aeruginosa from a contaminated freshwater marsh by using crude oil as its only carbon source. In addition, under both dynamic and static culture conditions, the isolated strain shown encouraging results in the degradation of important components of crude oil, including as n-alkanes, alkylcyclohexane, alkylbenzene, and alkyltoluene, in addition to several PAHs and aromatic chemical families. Since these wetlands are a major source of hydrocarbon-degrading bacteria, screening native bacteria with high degradation efficiency and the ability to break down a wide variety of crude oil components may provide a rich microbial resource for wetland restoration.25
Five different bacterial strains were found in soil samples from five different oil-polluted areas. The ability of the microorganisms to degrade diesel fuel was examined by researchers. By using phenotypic analysis, the bacterial colonies were shown to be Pseudomonas species, specifically P. putida, P. maltophilia, and P. mallei. They discovered a variety of bacteria, such as Acinetobacter lowffi and Enterobacter cloacae. Pseudomonas members were discovered to be the most common in oil-polluted Kuwaiti desert soil samples that were treated in several methods, along with Bacillus, Streptomyces, and Rhodococcus.28 Further as an add up scientific decipher, P. aeruginosa remains the subject of recent researchers e.g., evaluation of antibacterial activity of disinfectants,29 drug resistanxe30,31 and medical research tool.32,33
Pseudomonas aeruginosa is a kind of bacteria that can live in soil that has been polluted with oil products. The results of this experiment showed that the P. aeruginosa that was isolated could break down oil spills and can use in petroleum biodegradation. The P. aeruginosa isolates that were evaluated were very resistant to the medications that were utilized in this investigation.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- Arjoon K, Speight JG. Chemical and Physical Analysis of a Petroleum Hydrocarbon Contamination on a Soil Sample to Determine Its Natural Degradation Feasibility. Invention. 2020;5(3):43.
Crossref - Ajona M, Vasanthi P. Bioremediation of petroleum contaminated soils-A review. Mater Today Proc. 2021;45(7):7117-7122
Crossref - Nametkin, S.S. Khimiya Nefti; Izd-vo AN SSSR: Moscow, Russia. 1965
- Kumar P, Dey SR, Sharma M, Singh J. Microbes in bioremediation of petroleum pollutants. Development in Waste Water Treatment Research and Processes. 2025:557-580.
Crossref - Varjani SJ. Microbial degradation of petroleum hydrocarbons. Bioresour Technol. 2017;223:277-286.
Crossref - Fenibo EO, Selvarajan R, Abia ALK, Matambo T. Medium-chain alkane biodegradation and its link to some unifying attributes of alkB genes diversity. Sci Total Environ. 2023;877:162951.
Crossref - Yan G, Cai B, Chen C, et al. Bioremediation of crude oil contaminated soil. Pet Sci Technol. 2015;33(6):717-723.
Crossref - Atlas RM, Bartha R. Microbial Ecology: Fundamentals and Applications; Benjamin/Cummings Pub. Co.: San Francisco, CA, USA. 1982;70.
Crossref - Kumari S, Das S. Bacterial enzymatic degradation of recalcitrant organic pollutants: catabolic pathways and genetic regulations. Environ Sci Poll Res. 1982;30(33):79676-79705.
Crossref - Patel AB, Shaikh S, Jain KR, Desai C, Madamwar D. Polycyclic Aromatic Hydrocarbons: Sources, Toxicity, and Remediation Approaches. Front Microbiol. 2020;11:2675.
Crossref - Maddela NR, Scalvenzi L, Venkateswarlu K. Microbial degradation of total petroleum hydrocarbons in crude oil: A field-scale study at the low-land rainforest of Ecuador. Environ Technol. 2016;38(20):2543-2550.
Crossref - Su Q, Yu J, Fang K, et al. Microbial removal of petroleum hydrocarbons from contaminated soil under arsenic stress. Toxics. 2023;11(2):143.
Crossref - Bossert I, Bartha R. Fate of petroleum in soil ecosystems. In Petroleum Microbiology; Atlas, R.M, Ed, Macmillan: New York, NY, USA. 1984:435-473.
- Gouthami K, Mallikarjunaswamy AMM, Bhargava RN, et al. Microbial Biodegradation and Biotransformation of Petroleum Hydrocarbons: Progress, Prospects, and Challenges. Genomics Approach to Bioremediation: Principles, Tools, and Emerging Technologies. 2023:229-247.
Crossref - Chunyan X, Qaria MA, Qi X, Daochen Z. The role of microorganisms in petroleum degradation: Current development and prospects. Sci Total Environ. 2023;865:161112.
Crossref - Kassenova Z, Iskakov Y, Bolat Y, et al. Effectiveness of oil-contaminated soil reclamation with humic preparations. Int J Agri Biosci. 2024;13(3):474-487.
Crossref - Widdel F, Rabus R. Anaerobic biodegradation of saturated and aromatic hydrocarbons. Curr Opin Biotechnol. 2001;12(3):259-276.
Crossref - Chen L, Zheng X, Zhang K, et al. Sustained-release nitrate combined with microbial fuel cell: A novel strategy for PAHs and odor removal from sediment. J Hazard Mater. 2001;455:131610.
Crossref - Bayoumi RA, Abul-Hamd AT. Optimization of Bacterial Biodegradation of Toluene and Phenol Under Different Nutritional and Environmental Conditions. J Appl Sci Res. 2010;6(8):1086-1095.
- Aislabie J, Saul DJ, Foght JM. Bioremediation of hydrocarbon-contaminated polar soils. Extremophiles. 2006;10(3):171-179.
Crossref - Yang L, Li W, Liu J, et al. Nitrate-dependent ferrous oxidation: Feasibility, mechanism, and application prospects for wastewater treatment. J Water Process Eng. 2024;60:105226.
Crossref - Sabate J, Vinas M, Solanas AM. Laboratory-scale bioremediation experiments on hydrocarbon-contaminated soils. Int Biodeterior Biodegradation. 2004;54(1):19-25.
Crossref - Almansoory AF, Talal A, Al-yousif AN, Hazaimeh M. Isolation and identification of microbial species for hydrocarbon degradation in contaminated soil and water. Plant Archives. 2019;19(1):971-977
- Kridi N, Al-Shater MS, Al Zoubi MM. Isolation and identification of some bacterial isolates from soil contaminated with crude oil and Testing Their Effectiveness. Baghdad Sci J. 2021;18(4):1476-1484.
Crossref - Liu H, Yang G, Jia H, Sun B. Crude Oil Degradation by a Novel Strain Pseudomonas aeruginosa AQNU-1 Isolated from an Oil-Contaminated Lake Wetland. Processes. 2022;10(2):307.
Crossref - Kadivarian S, Rostamian M, Dashtbin S, et al. High burden of MDR, XDR, PDR, and MBL producing Gram negative bacteria causing infections in Kermanshah health centers during 2019-2020. Iran J Microbiol. 2023;15(3):359.
Crossref - Baban ST. Molecular detection of carbapenemase-producing Pseudomonas aeruginosa isolated from intensive care units of surgical specialty hospital in Erbil city. Med J Babylon. 2020;17(2):185-193.
Crossref - Saadoun I. Isolation and Characterization of Bacteria from Crude Petroleum Oil Contaminated Soil and Their Potential to Degrade Diesel Fuel. J Basic Microbiol. 2002;42(6):420-428.
Crossref - Ibrahim HM, Salem HM, Alamoudi SA, et al. Evaluating the bactericidal activity of various disinfectants against Pseudomonas aeruginosa contamination in broiler chicken hatcheries. Pak Vet J. 2024;44(3):683-690.
- Abdullah RM, Ali S, Aslam B, Arshad MI. Molecular characterization and drug resistance pattern of Pseudomonas aeruginosa isolated from poultry meat and meat products. Pak Vet J. 2024;44(3):812-818.
- Walaa GN, Ahmed LI, Awad AAN, Taher EM. Occurrence, Antimicrobial Resistance, and Virulence of Staphylococcus aureus, Escherichia coli, and Pseudomonas aeruginosa Isolated from Dairy Products. Int J Vet Sci. 2024;13(2): 218-225.
Crossref - Tharwat M, El-Magawry S, Kandeel A, Alkheraif AA. Mesenteric Abscessation caused by Pseudomonas aeruginosa in a Thoroughbred Mare: Clinical, Etiological, Hematobiochemical, Sonographic and Treatment Follow-up. Int J Vet Sci. 2024;13(6):1017-1022.
Crossref - Mohammed BQ, Abdullah AH, Rayshan AR. Pseudomonas that causes otitis in dogs: An increasing opposition. Int J Agri Biosci. 2024;13(1):59-64.
Crossref
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.