ISSN: 0973-7510
E-ISSN: 2581-690X
Bile salt hydrolase is an enzyme from bacteria that can hydrolyze bile acid to be insoluble form and this event lead to reducing of cholesterol level in serum. A new strain of lactic acid bacteria, which was isolated from fermented spider plant in local market of Thailand, showed ability to produce bile salt hydrolase and has potential to be probiotic bacteria with good colonizing properties. The new strain was identified as Lactobacillus plantarum and gene that responsible for bile salt hydrolase production is located on plasmid. From the study could be suggested that novel strain of LAB possible to be further used as cholesterol lowering probiotic bacteria and fermented spider plant would be cheap price cholesterol lowering foods for people in local area.
Bile salt, hydrolase, lactic acid bacteria, fermented spider plant
Spider plant (Cleome gynandra L.), which belongs to the Cleomaceae family, is plant that normally be found worldwide. In Asian country especially South-east Asia, this kind of plant can be cooked as a food 1. Because of fresh plant containing hydrocyanic acid, which is toxic for human at nervous system, cook by heating or fermenting can reduce toxicity of spider plant 2. Fermented spider plant is generally sold in local market with very cheap price. Unpublished data show that fermented spider plant is almost containing potential probiotic lactic acid bacteria.
Lactic acid bacteria (LAB) is the bacteria that can produce lactic acid as by-product. The genera of bacteria that commonly define as LAB are Lactobacillus sp., Streptococcus sp., Lactococcus sp., etc3. Almost of LAB could be assigned as probiotic bacteria which can provide benefit to the host 4, 5. Major criteria for probiotics designation are including with ability to survive and colonize in gastrointestinal tract of the host. In addition, probiotics should promote health status of host by any mechanism, e.g. immune modulation, vitamin production and bile acid deconjugation.
Bile is a natural emulsifying substance synthesised by hepatocytes using precursor such as cholesterol which is one of the cause for cardiovascular disease. In normal physiology, bile is conjugated by the liver and is transported through the enterohepatic system, where it emulsifies ingested fats for nutrient absorption. After use, it can be reabsorbed, recycled or excreted out. Bile circulating through the enterohepatic system undergoes various transitions in forms, which can be mediated by intestinal microflora 6. The bsh gene that encode to produce bile salt hydrolase were found in some strains of bacteria and this enzyme mediates the hydrolysis of conjugated bile acids to unconjugated form. From its function, bile salt hydrolase is believed to possess potential cholesterol-lowering effects. Hydrolysis action of bile salt hydrolase splits peptide bonds of bile salts, freeing the amino acid taurine and glycine from the bile salt molecule and inducing its precipitation. With decreased reabsorption ability, cholesterol precursors are being pulled for bile synthesis due to less bile salts recycling, resulting in the overall reduction of cholesterol content 7.
Because of there is no information for LAB isolates with ability to produce bile salt hydrolase from fermented spider plant, isolation and study various strains of LAB from fermented spider plant for bile salt hydrolase activity to use as a potential cholesterol-lowering probiotic bacteria are the aims of this study.
Microorganisms and growth condition
LAB isolates were grown with Mann, Rogosa and Sharpe (MRS) medium and cultivated at 37°C. For pathogenic test organisms, including Staphylococcus aureus, Enterococcus faecalis, Escherichia coli, Vibrio cholerae, Salmonella Typhimurium, Pseudomonas aeruginosa and Candida albicans, were cultured in Mueller-Hinton medium with 37°C incubation.
Screening of lactic acid bacteria from fermented spider plant
Fermented spider plants were collected from local market in Pathum Thani. Sample was serially diluted in normal saline solution (NSS) and spread on MRS agar plate containing 0.5% CaCO3. After incubation at 37°C for 48 – 72 h, bacterial colonies with clear zone were selected for determination of LAB properties.
Colonizing properties
The gastric fluid tolerance was done according the methods described by Mota et al8. To determine tolerance to the acid, the bacterial isolates were grown overnight in MRS medium at 37°C. The overnight culture of LAB was washed with NSS and adjusted the cell density to McFarland no. 0.5. A hundred microliters of adjusted bacterial suspension was added to 900 µl of sterile synthetic gastric juice (2 g of NaCl and 3.2 g of pepsin in 1 liter of distilled water, the pH was adjusted to 2.5 with concentrated HCl). The NSS was added instead of the gastric juice for the bacterial growth control tube. Both tubes, the test and control, were incubated at 37°C for 3 h. After incubation, the bacterial viability was determined by plate count technique on MRS agar and percentage of survival was calculated as (1-log cfuacid/log cfucontrol) x 100.
The bile salt tolerance was done according to method described by Walker and Gilliland 9. Briefly, the isolate was grown overnight in MRS broth and 1% of the inoculum was added to MRS containing 0.3% bile salt (Sigma-Aldrich, USA). The amount of bacteria was then determined by plate count technique before and after incubation the suspension at 37°C for 24 h. The percentage of bile salt tolerance was calculated as (1 – log after incubation/ O.D.log before incubation) x 100.
To evaluate the hydrophobicity of LAB cell surface, microbial adhesion to solvents (MATS) was determined in this study8 In brief, the 12 h cultures were collected by centrifugation, washed twice and adjusted with NSS to an O.D. of 0.4 – 0.5 (A0) at 600 nm. The 0.2 ml of xylene was added to 1.2 ml of bacterial suspension incubated at room temperature for 10 min and then mixed by vortexing for 2 min. The lower aqueous phase was collected after the tube was stood for 30 min and measured the O.D. at 600 nm (A1). The percentage of MATS was calculated as (1 – A1/A0) x 100.
Bile salt hydrolase activity and blood hemolysis
To determine activity of bile salt hydrolase, taurodeoxycholic acid (TDCA) (Sigma-Aldrich, USA) and glycodeoxycholic acid (GCDA) (Sigma-Aldrich, USA) were added in MRS as detective ingredient in the experiment. LAB isolates were cultured on MRS containing 0.5% of taurodeoxycholic acid or glycodeoxycholic acid and 0.37% CaCl2 then incubated at 37°C for 48 h. Bile salt hydrolase activity was evaluated by halozone production around colony on agar plate 10.
For blood hemolysis, the isolates were streak on blood agar. Blood hemolytic zone were observe after incubated at 37°C for 72 h.
Antimicrobial activity
The antimicrobial activity of the isolates was determined by well diffusion assay 11. The 20 ml of Mueller-Hinton agar (MHA) plate with a well (made from cork borer no.3) was spread with the indicator bacteria and filled in the well with 0.1 ml of supernatant culture, which had been adjust pH to 7 and sterilized with membrane filter, collected from LAB isolates. The MHA plate was incubated at 37°C for 24 h and measured for inhibition zone.
Antibiotic susceptibility
For antibiotic susceptibility, ampicillin, chloramphenicol, ciprofloxacin, erythromycin, nalidixic acid, penicillin, tetracycline and vancomycin were antibiotics in this study. The CLSI (Clinical and laboratory standards institute) guideline was followed as standard protocol for MIC (minimal inhibitory concentration) determination. Overnight culture of LAB isolates in MRS medium was adjusted turbidity equal to 0.5 MacFarland standards with normal saline solution. The adjusted bacterial suspension was diluted 100-fold with MRS broth and added to a 96 well plate filled with MRS containing a serial 2-fold diluted of each antibiotics. The MIC was observed and interpreted after incubated at 37°C for 24 h.
Bacterial identification
To identify LAB isolate, API 50 CHL (Biomeriux, France) was used and the 16s rRNA was analyzed sequence to confirm result of biochemical tests. Following API 50 CHL protocol, 2 days culture of LAB isolate on MRS agar was mixed with reagent of the kit and added into the well strip. Then filled the mixture in the well with oil immersion and incubated at 37°C. After 48 h incubation, the result was recorded and analyzed on https://apiweb.biomerieux.com.
For 16s rRNA sequence analysis, bacterial genomic DNA was extracted with GF-1 bacterial DNA extraction kit (Vivantis, Malaysia) and PCR product of 16s rRNA was amplified with F8 (5’-AGAGTTTGATCCTGGCTCAG-3’) and R1492 (5’-GGTTACCTTGTTACGACTT-3’) 12. Amplification was performed as following condition; 95°C for 10 min and 34 cycles of 95°C for 1 min, 55°C for 40 s, 72°C for 2 min and 72°C for 5 min. The PCR product was mixed with Novel Juice (GeneDireX, USA) and visualized on 0.8% agarose gel (Invitrogen, USA).
Bile salt hydrolase gene amplification
For bsh gene amplification, genomic DNA was used as a template and primer for PCR amplification were Lp1F (5’-CGCTACCGGAATTCATGT GTACTGCCATA-3’) and Lp975R (5’-CGCTACCCAAGCTTT TAGTTAACTGCATA-3’) 13. Amplification was performed as following condition; 95°C for 10 min and 34 cycles of 95°C for 1 min, 50°C for 40 s, 72°C for 2 min and 72°C for 5 min. Finally, the PCR product was visualized on 0.8% agarose gel (Invitrogen, USA).
Screening and isolation of bile salt hydrolase producing LAB
A hundred and thirty-nine isolates were obtained from isolation of LAB from fermented spider plant. After screening with basic characteristic of LAB, gram positive bacteria with catalase negative, 74 isolates were included in this study. From bile salt hydrolase activity determination with plate assay, 17 isolates of LAB (22.97% of isolates in this study) showed halozone around colony on MRS containing GDCA (figure 1) which refer to ability of bile salt hydrolase production. Otherwise, halozone was not observed on MRS containing TDCA plate. Bile salt hydrolase is an important enzyme in bile acid deconjugation and might be major characteristic to reveal before using of a novel probiotic isolate for human. Fermented foods are the source of LAB and the LAB isolates from this source commonly has ability to be probiotics. LAB with probiotic properties could be isolated from fermented spider plant in this study, this confirm the report that fermented spider plant is responsible for LAB with probiotic properties isolation 14. Bile salt hydrolase activity has been observed only on MRS agar plate containing GCDA but not on MRS with TDCA for all isolates, however glycine- to taurine- bile acid conjugates ratio is about 3.5:1 in adult human bile 15 so deconjugation activity for glycine conjugated bile salt would be noticeable to determine.
Fig. 1. Halozone around colony of bile salt hydrolase producing LAB after culture on MRS medium containing bile acid (GDCA)
Probiotic properties of LAB isolates
All isolates that can produce bile salt hydrolase were determined colonizing properties which important for probiotic designation. Colonizing properties including acid tolerance, bile salt tolerance and hydrophobicity of the isolates were examined 16. Bile can cause destruction of bacteria with surfactant properties leading to bacterial cell lysis 17 and probiotic bacteria should alive in this condition. Almost of bile salt hydrolase positive isolates have ability to survive in MRS containing 0.3% bile salt. The isolates that can survive in medium containing bile salt were determine ability to survive in simulated gastric juice and the result found that there are 3 isolates have this property. The pH of fermented foods normally are around 4 – 4.5 while pH of gastric condition is approximately 2 or rise to around 3 after ingestion with foods 18. Probiotic bacteria must be survived in this very low pH condition of stomach prior pass through to duodenum region which pH is about 7. Three isolates of LAB were tested for hydrophobicity with xylene to determine attachment ability to hydrocarbon which associated to cell adherent. Hydrophobicity of bacteria is related with ability to attach epithelium cells 19 and cut off of the test for bacterial isolates should be more than 50%8. Only SF21 isolates show strong hydrophobicity with 77.32%.
Safety of probiotic is also needed criteria to prove before use in human 20. In this study, blood hemolysis was evaluated and the SF21 isolate was not show ability to lyse red blood cell. Finally, SF21 isolate have ability to produce bile salt hydrolase with good colonizing properties but without blood hemolysis were obtained from the experiments with 77.32% of hydrophobicity, 32.45% survival in gastric juice and 87.30 % of bile salt tolerance. The SF21 was selected for further identification.
The SF21 isolate could not inhibit test organisms which are gram positive bacteria, gram negative bacteria and yeast. Antibiotic susceptibility test was determined and. SF21 isolate demonstrated sensitive to ampicillin, chloramphenicol, erythromycin, penicillin and tetracycline but resistance against ciprofloxacin, nalidixic acid and vancomycin. Because of antibiotic resistance gene horizontal gene transfer can cause problem and should be concerned21,20, sensitivity of SF21 to several antibiotics could be useful for probiotic using. According to resistance to vancomycin is one criteria for lactobacilli identification 22, this isolate might be a species in lactobacilli that would be confirmed in further experiment.
LAB isolates identification
The isolate has been identified by API 50 CHL test kit and 16s rRNA sequencing. Both results show that the SF21 isolate is L. plantarum with 99% identity. L. plantarum is a LAB that can be found generally in fermented foods and normally be used for health benefit. Various reports show that L. plantarum has been designated as probiotic bacteria with diverse properties, e.g. aflatoxin binding 23, antimicrobial activity 23,24 , antioxidant activity 25, bile salt hydrolase production and cholesterol removal 26, 27. From previous study, bile salt hydrolase of L. plantarum was evaluated in several strains 28-33. This could be suggested that L. plantarum SF21 probably be used as novel probiotic bacteria to reduce level of cholesterol in human serum.
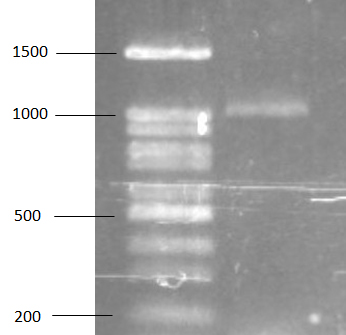
The bsh gene determination
For bsh gene, PCR was performed to detect bsh gene with Lp1F and Lp975R primer. Specific band approximately 1000 bp has been found on gel from electrophoresis (figure 2) and the PCR product was sent to determine nucleotide sequence. According to the result of sequencing, bsh gene of L. plamtarum SF21 was similar to bsh gene of L. plantarum B1 with 99% identity. For PCR amplification, PRC product of bsh gene was amplified from extracted plasmid and this result obviously revealed the location of bsh gene of L. plantarum SF21. Location of bsh gene has been previously reported that could be found on chromosome of E. faecium and other enterococci bacteria 34. In L. salivarius UCC118, the bacteria contain two genes of bile salt hydrolase that were originally located on both chromosome and megaplasmid 35. For location of bsh gene in L. plantarum, there is ambiguous report in previous works so this study would be one result that defined the bsh gene location in L. plantarum.
In summary, this is the first report for bile salt hydrolase producing lactic acid bacteria that can be isolated from local fermented spider plant. Possibility to find bile salt hydrolase producing LAB lead to conclude that fermented spider plant probably regard as cholesterol lowering foods with cheap price for people in local area such as South-east Asian countries. Finally, this strain of LAB need to be further studied for safety and cholesterol depletion in animal model to develop the isolate for using as potential probiotic bacteria and cholesterol lowering agent in human.
ACKNOWLEDGMENTS
We would like to thank Chulabhorn International College of Medicine for all facilities. The authors gratefully acknowledge the financial support provided by Thammasat University Research Fund under the TU New Research Scholar, contract no 25/2559.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
- Wu TH, Solberg SO, Yndgaard F, Chou YY. Morphological patterns in a world collection of Cleome gynandra. Genet Resour Crop Ev. 2018; 65(1):271-83.
- Onyango CM, Kunyanga CN, Ontita EG, Narla RD, Kimenju JW. Current status on production and utilization of spider plant (Cleome gynandra L.) an underutilized leafy vegetable in Kenya. Genet Resour Crop Ev. 2013; 60(7):2183-9.
- Nionelli L, Curri N, Curiel JA, Di Cagno R, Pontonio E, Cavoski I, et al. Exploitation of Albanian wheat cultivars: characterization of the flours and lactic acid bacteria microbiota, and selection of starters for sourdough fermentation. Food Microbiol. 2014; 44:96-107.
- Fuller R. Probiotics in man and animals. J Appl Bacteriol. 1989; 66(5):365-78.
- de Vrese M, Schrezenmeir J. Probiotics, prebiotics, and synbiotics. Adv Biochem Eng Biotechnol. 2008; 111:1-66.
- Kumar RS, Brannigan JA, Prabhune AA, Pundle AV, Dodson GG, Dodson EJ, et al. Structural and functional analysis of a conjugated bile salt hydrolase from Bifidobacterium longum reveals an evolutionary relationship with penicillin V acylase. J Biol Chem. 2006; 281(43):32516-25.
- Begley M, Hill C, Gahan CG. Bile salt hydrolase activity in probiotics. Applied and environmental microbiology. 2006; 72(3):1729-38.
- Mota RM, Moreira JL, Souza MR, Horta MF, Teixeira SM, Neumann E, et al. Genetic transformation of novel isolates of chicken Lactobacillus bearing probiotic features for expression of heterologous proteins: a tool to develop live oral vaccines. BMC biotechnology. 2006; 6:2.
- Walker DK, Gilliland SE. Relationship among bile tolerance, bile salt deconjugation, and assimilation of cholesterol by Lactobacillus acidophilus. J Dairy Sci. 1993; 76(4):956-61.
- Lim HJ, Kim SY, Lee WK. Isolation of cholesterol-lowering lactic acid bacteria from human intestine for probiotic use. Journal of veterinary science. 2004; 5(4):391-5.
- Panya M, Lulitanond V, Tangphatsornruang S, Namwat W, Wannasutta R, Suebwongsa N, et al. Sequencing and analysis of three plasmids from Lactobacillus casei TISTR1341 and development of plasmid-derived Escherichia coli–L. casei shuttle vectors. Applied microbiology and biotechnology. 2012; 93(1):261-72.
- Eden PA, Schmidt TM, Blakemore RP, Pace NR. Phylogenetic analysis of Aquaspirillum magnetotacticum using polymerase chain reaction-amplified 16S rRNA-specific DNA. International journal of systematic bacteriology. 1991; 41(2):324-5.
- Kumar R, Grover, S., Mohanty, AK. and Batish, VK. Molecular cloning and sequence analysis of bile salt hydrolase gene (bsh) from Lactobacillus plantarum MBUL90 strain of human origin. Food Biotech. 2010; 24(3):215-26
- Tanasupawat S, Komagata K. Lactic acid bacteria in fermented foods in Thailand. World J Microbiol Biotechnol. 1995; 11(3):253-6.
- Hardison WG, Grundy SM. Effect of bile acid conjugation pattern on bile acid metabolism in normal humans. Gastroenterology. 1983; 84(3):617-20.
- Dunne C, O’Mahony L, Murphy L, Thornton G, Morrissey D, O’Halloran S, et al. In vitro selection criteria for probiotic bacteria of human origin: correlation with in vivo findings. Am J Clin Nutr. 2001; 73(2 Suppl):386S-92S.
- Monte MJ, Marin JJ, Antelo A, Vazquez-Tato J. Bile acids: chemistry, physiology, and pathophysiology. World J Gastroenterol. 2009; 15(7):804-16.
- Carriere F, Moreau H, Raphel V, Laugier R, Benicourt C, Junien JL, et al. Purification and biochemical characterization of dog gastric lipase. Eur J Biochem. 1991; 202(1):75-83.
- Farrag HA, A AKE-D, Mohamed El-Sayed ZG, Abdel-Latifissa S, Kamal MM. Microbial colonization of irradiated pathogenic yeast to catheter surfaces: Relationship between adherence, cell surface hydrophobicity, biofilm formation and antifungal susceptibility. A scanning electron microscope analysis. Int J Radiat Biol. 2015; 91(6):519-27.
- Saarela M, Mogensen G, Fonden R, Matto J, Mattila-Sandholm T. Probiotic bacteria: safety, functional and technological properties. J Biotechnol. 2000; 84(3):197-215.
- Imperial IC, Ibana JA. Addressing the Antibiotic Resistance Problem with Probiotics: Reducing the Risk of Its Double-Edged Sword Effect. Front Microbiol. 2016; 7:1983.
- Devriese LA, Butaye P. Vancomycin susceptibility an an aid to the identification of lactobacilli. Lett Appl Microbiol. 1998; 27(2):121.
- Huang L, Duan C, Zhao Y, Gao L, Niu C, Xu J, et al. Reduction of Aflatoxin B1 Toxicity by Lactobacillus plantarum C88: A Potential Probiotic Strain Isolated from Chinese Traditional Fermented Food “Tofu”. PLoS One. 2017; 12(1):e0170109.
- Oldak A, Zielinska D, Rzepkowska A, Kolozyn-Krajewska D. Comparison of antibacterial activity of Lactobacillus plantarum strains isolated from two different kinds of regional cheeses from Poland: oscypek and korycinski cheese. Biomed Res Int. 2017; 2017:6820369.
- Li S, Zhao Y, Zhang L, Zhang X, Huang L, Li D, et al. Antioxidant activity of Lactobacillus plantarum strains isolated from traditional Chinese fermented foods. Food Chem. 2012; 135(3):1914-9.
- Fuentes MC, Lajo T, Carrion JM, Cune J. Cholesterol-lowering efficacy of Lactobacillus plantarum CECT 7527, 7528 and 7529 in hypercholesterolaemic adults. Br J Nutr. 2013; 109(10):1866-72.
- Costabile A, Buttarazzi I, Kolida S, Quercia S, Baldini J, Swann JR, et al. An in vivo assessment of the cholesterol-lowering efficacy of Lactobacillus plantarum ECGC 13110402 in normal to mildly hypercholesterolaemic adults. PLoS One. 2017; 12(12):e0187964.
- Duary RK, Batish VK, Grover S. Relative gene expression of bile salt hydrolase and surface proteins in two putative indigenous Lactobacillus plantarum strains under in vitro gut conditions. Mol Biol Rep. 2012; 39(3):2541-52.
- Gu XC, Luo XG, Wang CX, Ma DY, Wang Y, He YY, et al. Cloning and analysis of bile salt hydrolase genes from Lactobacillus plantarum CGMCC No. 8198. Biotechnol Lett. 2014; 36(5):975-83.
- Kumar R, Grover S, Batish VK. Hypocholesterolaemic effect of dietary inclusion of two putative probiotic bile salt hydrolase-producing Lactobacillus plantarum strains in Sprague-Dawley rats. Br J Nutr. 2011; 105(4):561-73.
- Kumar R, Grover S, Kaushik JK, Batish VK. IS30-related transposon mediated insertional inactivation of bile salt hydrolase (bsh1) gene of Lactobacillus plantarum strain Lp20. Microbiol Res. 2014; 169(7-8):553-60.
- Lambert JM, Bongers RS, de Vos WM, Kleerebezem M. Functional analysis of four bile salt hydrolase and penicillin acylase family members in Lactobacillus plantarum WCFS1. Applied and environmental microbiology. 2008; 74(15):4719-26.
- Ren J, Sun K, Wu Z, Yao J, Guo B. All 4 bile salt hydrolase proteins are responsible for the hydrolysis activity in Lactobacillus plantarum ST-III. J Food Sci. 2011; 76(9):M622-8.
- Wijaya A, Hermann A, Abriouel H, Specht I, Yousif NM, Holzapfel WH, et al. Cloning of the bile salt hydrolase (bsh) gene from Enterococcus faecium FAIR-E 345 and chromosomal location of bsh genes in food enterococci. J Food Prot. 2004; 67(12):2772-8.
- Fang F, Li Y, Bumann M, Raftis EJ, Casey PG, Cooney JC, et al. Allelic variation of bile salt hydrolase genes in Lactobacillus salivarius does not determine bile resistance levels. J Bacteriol. 2009; 191(18):5743-57.
© The Author(s) 2018. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.