ISSN: 0973-7510
E-ISSN: 2581-690X
Two whitish-colored gram-positive strains producing Polyhydroxyalkanoates (PHAs) were isolated from a soil sample from Nalgonda district in the Telangana state of India. With the help of Sudan black B staining, ten isolates with lipids, which showed bluish black color were selected from twenty-six bacterial strains which were selected randomly and purified from the serial diluted plate. Among ten isolates, 3D1 and 3D10 isolates were confirmed with Nile blue A and Nile Red staining for their PHA granules producing capacity. These two isolates grew optimally at a temperature of 37°C and a pH of 9. Furthermore, these strains were able to resist NaCl up to 10%, whereas, optimum NaCl required for the growth of 3D1 was 2%, but optimum NaCl required for the growth of 3D10 was shown to be 4%. PHAs produced by the two strains, 3D1 and 3D10, were extracted and quantified, which produced 68% PHA with a polymer concentration of 4.902 g/L and 61% PHA with a polymer concentration of 4.023 g/L, respectively. Biochemical, Morphological and Molecular characterization were performed on these two isolates. These two strains, 3D1 and 3D10, were closely related to Bacillus sonorensis with similarity of 99.51% and Bacillus safensis subsp. safensis with similarity of 99.66%, respectively. The 16S rDNA gene sequences of these two isolates were submitted to the NCBI Gene bank and the accession numbers were also sought.
Sudan black B and Nile Red Staining, Morphological and Molecular Characterization, Nalgonda District
Polyhydroxyalkanoates (PHAs)/Bioplastics are the biodegradable, biocompatible and environmentally friendly storage materials, produced by bacteria, archea and few eukaryotes like yeasts and fungi. Polyhydroxyalkanoates are produced by microbes when they are supplied with high carbon source and other nutrients such as, nitrogen, oxygen, sulfur, or phosphorus are in limited conditions. Some of the bacteria which produce these PHAs are Alcaligenes latus, Ralstoniaeutropha, Bacillus megaterium, Pseudomonas oleovorans and Azotobacter beijerinckii etc. Microbial species from over 90 genera are reported to produce and accumulate these polyesters.1 Nearly 150 different hydroxyalkanoic acids are included into polyhydroxyalkanoates,2 One of the PHA family member i.e.polyhydroxybutyrate (PHB) is considered as important biodegradable polymer and best alternative to synthetic plastics, which are mostly non-degradable. The PHAs are generally accumulated as inclusions inside the intracellular membrane around 90% of the microorganisms’ cell dry weight. Similar to conventional plastics, which are derived from petroleum products (eg. polypropylene or polyethylene), these biodegradable polymers have similar mechanical properties. But, these are not like petrochemical based plastics which take several decades to undergo full degradation process. PHAs can be degraded quickly by a variety of microorganisms into CO2 and water. Plastic materials are causing serious environmental problems. Hence, two solutions were chosen to avoid the same. First one is recycling of use-plastic materials and second one is using reduced amount of plastic in the use of biodegradable plastics.3,4
These polymers have utilization in packaging, agriculture, and medicine, as they are immunologically compatible with human tissue5 and can be used as biofuel.6 Despite of these many important features, PHA production at industrial scale is not established.
PHAs are a good substitute for plastics made from oil, but they are still far from being able to compete with them, mostly because of their high manufacturing costs and lack of specialized rules.7 To commercialize PHA, more efforts were made to decrease the cost of production by developing different bacterial strains and more efficient production/purification process.8,9 As more than 50% of production cost 10, 11 is substrate itself12,13, using inexpensive substrate, waste material like cardboard industry effluent14, the use of waste and wastewater as feedstock for the PHA production,15 treating paper-mill wastewater16 and renewable substrates as source of nutrients source for bacteria for production of PHA can reduce the product cost of PHAs.17,18 AIK7 isolate could reach 35% CDW of PHA in 72 hours when grown in 10g/L waste glycerol whereas, 33% CDW of PHA in 120 hours with pure glycerol19. To increase the purity, one of the option, where Halophiles, which require high salinity for their growth can prevent microbial contamination and high intracellular osmotic pressure, which could help in increasing the PHA recovery rate. Halophiles have the capacity to use wide range of low-cost substrates also,20 like normal genetically modified organisms have the capacity to produce and accumulate more PHA than their wild strains,21 engineered extremophiles could produce more amount of PHA using carbon substrates. The PHA production capacity of the strain, Pseudomonas sp. strain-P in by utilizing of various cost-effective carbon substrates like rice bran, dates, and soy molasses was 90.9%, 82.6%, and 91.6%, respectively.22 The accumulation of PHB and its co-polymers was also observed in Bacillus and Pseudomomas sp. strain 012 and 045.23
During the 1950s, first attempt for commercial production was made by North-American Company W.R. Grace Co. But, due to efficiency issues in both production and purification it was not successful. Later, Imperial Chemical Industries (UK) began producing commercial PHAs in the name of Biopol, by using a mutant strain Cupriavidusnecator, NCIB 11599 in the presence of different carbon sources, including 1,4-butanediol, butyrolactone and 1,6-hexanediol.
The patents have been then transferred first to Zeneca, then to Monsanto, which are at present with Metabolix, Inc. (USA). At commercial scale, different bio-polyester products with different monomer compositions are produced with trade names such as, Nodax [poly(3HB-co-3HV)], Biogreen [poly (3-hydroxybutyrate-co-3-hydroxyalkanoate) poly (3HB-co-3HA)] and Biomer [poly(3HB)].10
This study is aimed to isolate, purify PHA producing isolates from soil sample of Nalgonda district and their characterization.
Sudan black B staining of bacterial isolates
Soil sample was obtained near petrol pump, Anneparthy village, Nalgonda district, Telangana. Serial dilution of 1g soil sample in 99ml sterile distilled water was performed.22 Later, 1 ml of different dilutions (10-4 to 10-6) were spread on a nutrient medium made of peptone (5 g/L), beef extract (3 g/L), sodium chloride (0.8 g/L), and agar (15 g/L). The inoculation plates were incubated at 28-37°C for 24-48 hours. To create pure cultures, the colonies were further subcultured. The pure cultures were then kept at 4°C on nutrient agar slants for use in subsequent analyses.24 To detect the lipophilic compounds of bacteria, 0.02% alcoholic Sudan black B solution was used for staining bacterial colonies. The plates were kept at room temperature for 30 min. After decanting excess dye, colonies were checked for lipophilic compound producers with bluish black colour and non-lipophilic compound producers with white colour.25
Nile blue A and Nile Red staining for PHA Producing Bacteria
The isolates which were showing positive results for Sudan black B staining were further checked for PHA production by Nile blue A and Nile Red staining, which are more specific stains for Polyhydroxyalkanoic acids (PHAs).26,27 The isolates grown for 48 hrs at RT in the presence of Nile Red dye in carbon rich nutrient agar medium composing of glucose 1% w/v,, beef extract 0.3%, peptone 0.5%, sodium chloride 0.8%, and agar 1.5% Nile Red at a concentrations of 0.5µg/mL, which didn’t inhibit the growth of the bacteria.27 The PHA accumulating bacterial colonies showed orange fluorescence when exposed under UV light (365nm) and intensity of their fluorescence is directly proportional to the PHA content inside the cells. The bacterial isolates with bright orange fluorescence were selected as PHA producers.
Morphological and Biochemical Characterization for Identification of Bacteria Producing PHA
Olympus Microscope was used to observe the morphology of two bacteria, 3D1 and 3D10 grown on nutrient agar. Other growth characteristics including colony colour and texture, cell shape were observed. Gram-staining of the bacteria was performed to differentiate between Gram positive and negative bacteria to characterize the 3D1 and 3D10 isolates.28 Spectrophotometer 104 (Systronics) was used to measure the growth of the 3D1 and 3D10 isolates. Different biochemical tests like Catalase, Oxidase, Methy Red test and Nitrate reduction were performed on the two isolates, 3D1 and 3D10, for their Biochemical characterization.29
The carbohydrate utilization tests were also performed on the two isolates, 3D1 and 3D10, using Hi-carbo Kit (https://himedialabs.com/TD/KB009.pdf) After 48h of growth at 37°C the results were interpreted based on the colours produced.
Molecular identification
16S rRNA gene amplification and analysis of PCR products
DNA was extracted from 3D1 and 3D10 strains separately after incubating bacterial colonies at 95°C for 10 minutes in thermal cycler. These were used as DNA templates in PCR reactions to amplify the 16s rRNA genes of 3D1 and 3D10 strains. DNA samples were stored at −20°C until use. In these PCR reactions, Universal primers used were 27f (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492r (5′-CGGYTACCTTGTTACGACTT-3′).30 PCR programme used for both 3D1 and 3D10 strains was: 94°C for 5min, 94°C for 30S, 54°C for 30S,72°C for 2min and 72°C for 5min. After PCR reactions, Agarose gel electrophoresis was performed on 2% agarose gel to analyze the PCR products of both 3D1 and 3D10 strains.
PCR products sequencing of 3D1 and 3D10 strains and their analysis
Purification of PCR products and 16s rDNA sequencing of both 3D1 and 3D10 strains were out sourced to analyse the 16s rRNA genes. Universal primers were used in this sequencing process. After getting PCR product sequences, they were subjected to Ez-Biocloud analysis (https://www.ezbiocloud.net/apps) to search for closely related type species and to identify them (isolates 3D1 and 3D10). Later on, phylogenetic trees of both 3D1 and 3D10 strains were constructed by using MEGA 11.0 phylogenetic tree analysis software.31
Extraction and Quantitative Analysis of PHA
The polymer production by the two isolates, 3D1 and 3D10 was done in 250 mL flasks containing 100 mL of E2 medium with 2% glucose as production medium. After 72 h of growth at 37°C and pH 7, 50ml culture broths were centrifuged at 10,000 rpm for 10 min. The two bacterial cell pellets of 3D1 and 3D10 separately each with 10 mL sodium hypochlorite solution was incubated at 50°C for 1 h to lyse the cells. The cell extracts obtained were further centrifuged at 15,000 rpm for 30 min and then cell pellet was washed sequentially with distilled water, acetone, followed by absolute ethanol. The pellet was then dissolved in chloroform and incubated overnight at 50°C in an incubator, further it was evaporated at room temperature to obtain a white residue.32
For quantitative analysis, remaining 50ml cell cultures were used and cell pellets of both the isolates, 3D1 and 3D10 were dried to estimate the dry cell weights (DCW) in units of g/L Residual biomasses of the both isolates were estimated by calculating the difference between DCW and dry weight of extracted PHA.25 This was calculated to determine the cellular weight and accumulations of any other polymers other than PHAs. The intracellular PHA accumulation is calculated with the help of the dry weight of extracted PHA present in the dry cell weight, where:
Residual Biomass (g/L) = DCW (g/L)-Dry weight of extracted PHA (g/L)
% of PHA accumulation = Dry weight of extracted PHA (g/L) x100%/ DCW (g/L)
Isolation and Selection of PHA Producers
Serial dilution of soil sample, plating, purification of strains led to the isolation of twenty six bacteria. Out of which only ten isolates showed black-blue coloration after Sudan-Black staining, indicating presence of lipophilic compounds. Among those ten isolates, two organisms, 3D1 and 3D10 showed positive result with Nile blue A stain on the bacterial colonies (Figure 1) and Nile Red stain in the medium (not shown here) a specific dye for the direct screening of PHA accumulating bacteria.
Figure 1. PHA producing strains,3D1 and 3D10 showing pink/orange fluorescence under UV light with Nile blue A staining
Morphological and Biochemical Characterization for Identification of PHA Producers
The growth characteristics of two isolates, 3D1 and 3D10 were shown in Table 1. Growth curves of PHA producing strains, 3D1 and 3D10 in the NA medium were mentioned in Figures S1 and S2. Biochemical tests like Methyl red test, Voges-Proskauer test and Oxidase test showed negative, whereas, Catalase and Nitrate reduction tests were positive by the 3D1 and 3D10 organisms (Table 2). Carbohydrate utilization tests results of the two isolates, 3D1 and 3D10, performed using Hi-carbo Kit, after 48 h of growth at 37°C, were mentioned in Table 3.
Table (1):
The growth characteristics of two PHA producing isolates, 3D1 and 3D10.
No. |
Morphological character |
3D1 |
3D10 |
|---|---|---|---|
1 |
Colony colour |
Off white |
Off white |
2 |
Colony texture |
rough |
smooth |
3 |
Cell shape |
Rod |
Oval |
4 |
Gram staining |
G +ve |
G +ve |
Table (2):
Biochemical tests performance of PHA producing isolates, 3D1 and 3D10.
No. |
Biochemical test |
3D1 |
3D10 |
|---|---|---|---|
1 |
Methyl Red production |
-ve |
-ve |
2 |
Voges-Proskauer |
-ve |
-ve |
3 |
oxidase test |
-ve |
-ve |
4 |
Catalase test |
+ve |
+ve |
5 |
Nitrate reduction |
+ve |
+ve |
Table (3):
Carbohydrate utilization tests by two PHA producing isolates, 3D1 and 3D10.
No |
Carbohydrate utilization |
Result of 3D1 |
Result of 3D10 |
|---|---|---|---|
1 |
Lactose |
– |
– |
2 |
Xylose |
– |
– |
3 |
Maltose |
+ |
– |
4 |
Fructose |
+ |
+ |
5 |
Dextrose |
+ |
+ |
6 |
Galactose |
+ |
– |
7 |
Raffinose |
+ |
– |
8 |
Trehalose |
+ |
+ |
9 |
Melibiose |
– |
– |
10 |
Sucrose |
+ |
+ |
11 |
L-Arabinose |
+ |
+ |
12 |
Mannose |
+ |
– |
13 |
Inulin |
+ |
+ |
14 |
Sodium gluconate |
+ |
+ |
15 |
Glycerol |
– |
– |
16 |
Salicin |
+ |
+ |
17 |
Dulcitol |
– |
– |
18 |
Inositol |
+ |
+ |
19 |
Sorbitol |
+ |
+ |
20 |
Mannitol |
+ |
+ |
21 |
Adonitol |
– |
– |
22 |
Arabitol |
– |
– |
23 |
Erythritol |
– |
– |
24 |
α-Methyl-D-glucoside |
+ |
+ |
25 |
Rhamnose |
– |
– |
26 |
Cellobiose |
+ |
– |
27 |
Melizitose |
– |
– |
28 |
α-Methyl-D-Mannoside |
– |
– |
29 |
Xylitol |
– |
– |
30 |
ONPG |
– |
+ |
31 |
Esculin |
+ |
+ |
32 |
D-Arabinose |
+ |
– |
33 |
Citrate |
– |
+ |
34 |
Malonate |
– |
+ |
35 |
Sorbose |
– |
– |
Note: + = positive; – = negative.
Molecular identification
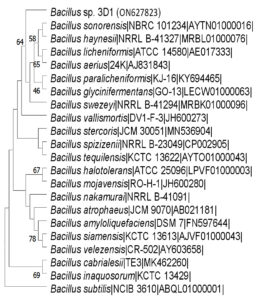
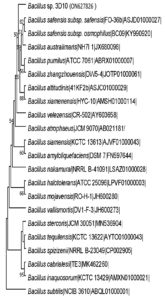
PCR products of the two isolates 3D1 and 3D10 were subjected to sequencing for their 16s rRNA gene analysis. The 16S rRNA gene sequences of two isolates (3D1 and 3D10) were submitted to EZBiocloud portal for analysis. The results showed that these two bacteria (3D1 and 3D10) were closely related to Bacillus sonorensis with similarity of 99.51% and Bacillus safensis subsp. safensis with similarity of 99.66% respectively. The 16S rDNA gene sequences of these two isolates 3D1 and 3D10 were submitted to the NCBI Gene bank and the accession numbers provided to these strains were ON627823 and ON627826, respectively.
Phylogenetic trees of both 3D1 and 3D10 strains constructed using MEGA 11.0 phylogenetic tree analysis software and were shown in Figure 2 and Figure 3.
Extraction and Quantitative Analysis of Polymer
Quantitative analysis of PHAs was performed after the extraction of polymers from the two isolates, 3D1 and 3D10. Isolate, 3D1 showed 7.2 g/L CDW with 68% PHA. Whereas, 3D10 showed 6.5 g/L CDW with 61% PHA. Metabolic processes are affected by pH changes, for example, 4.492 g/L PHB content was obtained by isolate NA10 at pH 7.5 whereas, Shivakumar33, reported pH 6.8 to 8.0 as optimum for PHB production by A.eutrophus.
Likewise, temperature changes beyond 35°C has negative effect on PHB production. According to the Bhuwal AK,14 report, up to 72h the production of PHB was increased and later it got reduced, which may be due to reduction in micronutrients and increase in metabolites which might cause negative effect on the PHB production. But according to Khanna S, Srivastava A34, Page WJ et al.35; Kluttermann K et al.,36 highest PHB production was achieved after 96h of incubation. Presence of different substrates in the media has effect on the production of PHA, for example, H. daqingensis showed DCW of 0.362+/_ 0.001g and produced 0.236 +/_ 0.003g PHA in the presence of 3% Algal biodiesel waste residue as sole substrate at 35⁰C with 5% NaCl in the fermenter with 0.5 vvm air flow rate and 200rpm for 48hrs.37
There is evidence that the PHA-polymer characteristics may be impacted by the extraction process.38 There are still no effective, safe, affordable, or environmentally friendly PHA extraction techniques available.
Two PHA producing strains, 3D1 and 3D10 which produced 68% PHA with polymer concentration of 4.902 g/L and 61% PHA with polymer concentration of 4.023 g/L respectively were isolated and characterized. These two PHA producing strains, 3D1 and 3D10 were closely related to Bacillus sonorensis with similarity of 99.51% and Bacillus safensis subsp. safensis with similarity of 99.66% respectively. To decrease the PHA production cost from these two PHA producing strains, 3D1 and 3D10, various cheap carbon sources can be substituted in the media as carbon source. Optimization of pH, temperature and incubation period could also be studied to improve the production of PHA by these wild strains.
Additional file: Additional Figure S1 and S2.
ACKNOWLEDGMENTS
The authors would like to thank Administrators of Mahatma Gandhi University, Nalgonda, India for providing the facilities for this research.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
MT designed the experiments. PSP, ESK and MT performed the experiments. MT and SVR wrote the manuscript. SVR reviewed and edited the manuscript for final publication.
FUNDING
None.
DATA AVAILABILITY
The 16S gene sequences from this work generated and/or analysed during the current study are deposited in the public data bank at NCBI database repository, Accession number ON627823 and ON627826.
ETHICS STATEMENT
This article does not contain any studies on human participants or animals performed by any of the authors.
- Zinn M, Witholt B, Egli T. Occurrence, synthesis and medical application of bacterial polyhydroxyalkanoate. Adv Drug Deliv Rev. 2001;53(1):5-21.
Crossref - Steinbuchel A.Perspectives for biotechnological production and utilization of biopolymers: metabolic engineering of polyhydroxyalkanoats biosynethesis pathways as a successful example. Macromol Biosci. 2001;1(1):1-24.
Crossref - Byrom D. Polymer synthesis by microorganisms: technology and economics. Trends Biotechnol. 1987;5(9):246-250.
Crossref - Lee SY, Chang HN. Production of poly(3-hydroxybutyric acid) by recombinant Escherichia coli strains: genetic and fermentation studies. Can J Microbiol. 1995;41(Suppl 1):207-215.
Crossref - Hazer B, Steinbuchel A. Increased diversification of polyhydroxyalkanoates by modification reactions for industrial and medical applications. Appl Microbiol Biotechnol. 2007;74 (1):1-12.
Crossref - Gao X, Chen J, Wu Q, Chen G. Polyhydroxyalkanoates as a source of chemicals, polymers, and biofuels. Curr Opin Biotechnol. 2011;22(6):768-774.
Crossref - Rosenboom JG, Langer R, Traverso G. Bioplastics for a circular economy. Nat Rev Mater. 2022;7(2):117-137.
Crossref - Tamer IM, Moo-Young M, Chisti Y. Optimization of poly(β-hydroxybutyric acid) recovery from Alcaligenes latus: combined mechanical and chemical treatments. Bioprocess Eng. 1998;19(6):459-468.
Crossref - Grothe E, Moo-Young M, Chisti Y. Fermentation optimization for the production of poly(β-hydroxybutyric acid) microbial thermoplastic. Enzyme Microbial Technol. 1999;25(1-2):132-141.
Crossref - Rudnik E. Compostable Polymer Materials. Elsevier Publishing, Amsterdam. 2008;28-31:183-198.
- Halami PM. Production of polyhydroxyalkanoate from starch by the native isolate Bacillus cereus CFR06. World J Microbiol Biotechnol. 2008;24(6):805-812.
Crossref - Yamane T. Cultivation engineering of microbial bioplastics production. FEMS Microbiol Rev. 1992;103(2-4):257-264.
Crossref - Yamane T. Yield of poly-D(−)-3-hydroxybutyrate from various carbon sources: a theoretical study. Biotechnol Bioeng. 1993;41(1):165-170.
Crossref - Bhuwal AK, Singh G, Aggarwal NK, Goyal V, Yadav A. Poly-β-hydroxybutyrate production and management of cardboard industry effluent by new Bacillus sp. NA10. Bioresour Bioprocess. 2014;1:9.
Crossref - Koller M, Marsalek L, de Sousa Dias MM, Braunegg G. Producing microbial polyhydroxyalkanoate (PHA) biopolyesters in a sustainable manner. N Biotechnol. 2017; 37(Pt A):24-38.
Crossref - Tamis J, Mulders M, Dijkman H, Rozendal R, Loosdrecht MCMv, Kleerebezem R. Pilot-scale polyhydroxyalkanoate production from paper mill wastewater: process characteristics and identification of bottlenecks for full-scale implementation. J Environ Eng. 2018;144.
Crossref - Choi J, Lee SY. Process analysis and economic evaluation for poly (3-hydroxybutyrate) production by fermentation. Bioprocess Eng. 1997;17(6):335-342.
Crossref - Castilho LR, Mitchell DA, Freire DMG. Production of polyhydroxyalkanoates (PHAs) from waste materials and by-products by submerged and solid-state fermentation. Bioresource Technol. 2009;100(23):5996-6009.
Crossref - Teeka J, Imai T, Cheng X, et al. Screening of PHA producing bacteria using biodiesel-derived waste glycerol as a sole carbon source. J Water Env Technol. 2010;8(4):371-381.
Crossref - Mitra R, Xu T, Xiang H, Han J. Current developments on polyhydroxyalkanoates synthesis by using halophiles as a promising cell factory. Microb Cell Fact. 2020;19(1):86.
Crossref - Chen G-Q, Jiang X-R. Engineering bacteria for enhanced olyhydroxyalkanoates (PHA) biosynthesis. Synthetic Sys Biotechnol. 2017;2(3):192-197.
Crossref - Aljuraifani AA, Berekaa MM Ghazwani AA. Bacterial biopolymer (polyhydroxyalkanoate) production from low-cost sustainable sources. Microbiol Open. 2019.8(6):e755.
Crossref - Sangkharak K, Prasertsan P. Screening and identification of polyhydroxyalkanoates producing bacteria and biochemical characterization of their possible application. J Gen Appl Microbiol. 2012;58(3):173-82.
Crossref - Gulab S, Anish K, Arpana M, Anita Y, Neeraj KA. Poly β-Hydroxybutyrate Production by Bacillus subtilis NG220 Using Sugar Industry Waste Water. BioMed R Inter. 2013;2013:952641.
Crossref - Bhuwal AK, Singh G, Aggarwal NK, Goyal V, Yadav A. Isolation and screening of polyhydroxyalkanoates producing bacteria from pulp, paper, and cardboard industry wastes. Int J Biomater. 2013;2013:752821.
Crossref - Law J, Slepecky RA. Assay of poly-beta-hydroxybutyric acid. J Bacteriol. 1961;82(1):33-36.
Crossref - Spiekermann P, Rehm BHA, Kalscheuer R, Baumeister D, Steinbuchel A. A sensitive, viable-colony staining method using Nile red for direct screening of bacteria that accumulate polyhydroxyalkanoic acids and other lipid storage compounds. Arch Microbiol. 1999;171(2):73-80.
Crossref - Vega-Vidaurri JA, Hernandez-Rosas F, Rios-Corripio MA, et al. Coproduction of polyhydroxyalkanoates and exopolysaccharide by submerged fermentation using autochthonous bacterial strains. Chem Pap. 2022;76:2419-2429.
Crossref - Shoaib M, Muzammil I, Hammad M, Bhutta ZA, Yaseen I. A Mini-Review on Commonly used Biochemical Tests for Identification of Bacteria. Int J Research Pub. 2020;54(1).
Crossref - Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173(2):697-703.
Crossref - Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA 6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725-2729.
Crossref - Singh G, Mittal A, Kumari A, Goel V, Aggarwal NK, Yadav A. Optimization of poly-B-hydroxybutyrate production from Bacillus species. European J Biol Sci. 2011;3(4):112-116.
- Shivakumar S. Optimization of process parameters for maximum poly-β-hydroxybutyrate production by Bacillus thuringiensis IAM 12077. Pol J Microbiol 2009; 58(2):149-154.
- Khanna S, Srivastava A. Recent advances in microbial polyhydroxyalkanoates. Process Biochem. 2005;40(2):607-619.
Crossref - Page WJ, Manchak J, Rudy B. Formation of poly(hydroxybutyrate-co- hydroxyvalerate) by Azotobacter vinelandii UWD. Appl Environ Microbiol. 1992;58(9):28-66.
Crossref - Kluttermann K, Tauchert H, Kleber HP. Synthesis of poly-beta-hydroxybutyrte by Agrobacterium radiobacter after growth on D-Carnitine. Acta Biotehnol. 2002;22(3-4):261-269.
Crossref - Dubey S, Mishra S. Efficient Production of Polyhydroxyalkanoate Through Halophilic Bacteria Utilizing Algal Biodiesel Waste Residue. Front Bioengg Biotechnol. 2021;9:624859.
Crossref - Palmieri S, Tittarelli F, Sabbatini S, et al. Effects of different pre-treatments on the properties of polyhydroxyalkanoates extracted from sidestreams of a municipal wastewater treatment plant. Sci Total Environ. 2021;801:149633.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.