ISSN: 0973-7510
E-ISSN: 2581-690X
This study aimed to assess and manage bacterial contamination in multiple batches of mesenchymal stem cell (MSC) cultures derived from rabbit bone marrow. Routine visual inspection and microscopic examination were employed for the detection of the contaminated cultures. The contaminated cultures were inoculated on Nutrient agar and multiple isolated colonies were subjected to Gram staining and biochemical characterization. Further, molecular identification of the bacterial isolates was performed using polymerase chain reaction. The determination of antibiotic susceptibility patterns was conducted using the Kirby-Bauer disc diffusion method. Among the 351 mesenchymal stem cell culture (SCC) flasks monitored, only 1.42% were found to be contaminated. Based on the phenotypic and biochemical characterization, the major bacterial contaminants were identified as Staphylococcus aureus, Bacillus spp., and Escherichia coli infiltrating during various stages of cell processing. Antibiotic susceptibility patterns revealed varying responses among isolates, crucial for effective antimicrobial strategies and maintaining aseptic conditions in SCCs. The study emphasizes the importance of regular monitoring to maintain sterile environments, validate culture quality, and uphold safety standards. The findings indicate the need to establish stringent quality control measures, crucial for the successful translation of MSC research into clinical applications. The research advocates for continuous monitoring, adherence to SOPs, and further investigations into preventive strategies for ensuring the safety and efficacy of MSC-based therapies.
Cell Culture Contamination, Stem Cell Culture, Mesenchymal Stem Cells, Bacterial Contaminants
Cell-based therapies represent a leading-edge approach for treating once-considered incurable ailments, with mesenchymal stem cells (MSCs) gaining widespread recognition in this field.1,2 Renowned for their capacity to differentiate into various cell types, MSCs contribute to tissue regeneration and repair.3,4 One of the primary therapeutic applications involves their use in regenerative medicine, particularly in the repair of damaged tissues such as bone, cartilage, and muscle.5-7 MSCs exhibit immunomodulatory effects, making them valuable in treating inflammatory and autoimmune conditions.2,8 However, the practical application of these therapies worldwide is hindered by biosafety concerns. Quality assurance in stem cell laboratories is pivotal for ensuring the safety, consistency, and efficacy of cell-based therapies.9 It encompasses a meticulous framework of standardized protocols, stringent monitoring, and rigorous documentation.10 Maintaining a meticulously monitored stem cell lab to check for potential pathogens is crucial.11 Detecting contamination is a complex process, and the ongoing debate revolves around the clinical consequences of infusing contaminated stem cells.
Contamination risks during cell harvest or subsequent manipulation in stem cell laboratories are a critical concern. Bacterial (including Mycoplasma), yeast, and fungal contaminations pose significant risks, potentially occurring during cell harvest or subsequent manipulation.12 These contaminants can compromise the integrity and safety of the final cell-based product. Cell harvesting procedures, especially from biological sources, demand stringent aseptic techniques to prevent microbial ingress.13 Subsequent manipulation steps, including expansion, differentiation, or genetic modification, also pose risks, as each handling instance creates a window for potential contamination.14 Maintaining controlled environments, implementing strict hygiene measures, and routinely monitoring cultures for any signs of microbial presence are essential safeguards against contamination.12 Even minute traces of unwanted microorganisms can profoundly impact the therapeutic potential and safety of stem cell products, emphasizing the critical need for robust contamination prevention strategies throughout the cell processing workflow.15
Regular microbiological checks on stem cell lines and operating within controlled environments are standard practices in current stem cell banks. These measures significantly lower the risk of contamination in the final product, adhering to good practices in the field.12 As part of our routine microbial surveillance, a comprehensive study was undertaken to identify prevalent contaminants within our stem cell lab over the past three years (from 2020 to 2022). This report specifically focuses on the isolation and identification of common contaminants found in SCCs derived from rabbit bone marrow mesenchymal stem cells (R-BMSCs) during this period. Our findings shed light on the specific issues faced in maintaining the quality of SCCs, fostering continuous improvement and adherence to best practices in stem cell research and bioprocessing.
Rabbit bone marrow-derived mesenchymal stem cell culture
Bone marrow-derived mesenchymal stem cells (BMSCs) were isolated and cultured from the bone marrow of New Zealand White rabbit as per the standard protocol.3,4 The rabbits were anesthetized, and the posterior iliac crest was aseptically exposed for bone marrow harvesting. Bone marrow aspirates were collected and immediately transferred to sterile tubes containing heparin anticoagulant. For BMSC isolation, the bone marrow aspirates were diluted with Dulbecco’s Modified Eagle Medium (DMEM) and subjected to density gradient centrifugation to separate the mononuclear cell fraction. The mononuclear cells were then carefully layered onto a density gradient medium, allowing for the isolation of BMSCs. The isolated cells were suspended in a culture medium composed of DMEM supplemented with fetal bovine serum (FBS) and antibiotics.3,4 In the primary culture establishment, the obtained BMSCs were seeded in tissue culture flasks and maintained in a humidified atmosphere with 5% CO2 at 37°C. The culture medium was changed every 2–3 days to eliminate non-adherent cells. Upon reaching confluence, primary BMSCs were trypsinized and subcultured. Subsequent subculture and expansion were performed by detaching cells with trypsin-EDTA and replating at an appropriate density.
Microbiological processing of the samples
Multiple batches of BMSC cultures were monitored for potential bacterial contamination. Detection of contaminated cultures was performed through routine microscopic examination and visual inspection for signs of contamination. The contaminated flasks were sampled for isolating the microorganisms. Aseptic techniques were employed during sample collection to avoid external contamination. A volume of 5-10 mL of sample media was collected for microbial isolation.
Isolation of microorganisms
For preliminary isolation appropriately diluted media samples were plated onto nutrient agar plates which were then incubated at 37°C for 24 h. After incubation, the pure cultures of the most frequently encountered bacterial contaminants were prepared and used for further testing. Each of the isolates was observed for colony characteristics like size, shape, and colour, and microscopic characteristics like Gram staining, shape, and arrangement of cells.
Gram staining and biochemical identification of the microorganisms
Multiple isolated colonies from the nutrient agar plates were stained using Gram’s staining method. The presumptive colonies were streaked onto different selective media to obtain the pure cultures. This was followed by the biochemical characterization using the catalase test, coagulase test [to differentiate Staphylococcus aureus (positive) from coagulase-negative Staphylococci], oxidase test, indole test, methyl red (MR) test, Voges Proskauer (VP) test, Simon Citrate agar test, Triple Sugar Iron (TSI) test and urea hydrolysis (urease test). For identification of Bacillus spp., all presumptive colonies, in addition to the above-mentioned tests, were subjected to endospore staining and nitrate reduction test as described previously.16
Molecular identification of the bacterial isolates
Isolation of DNA was done from broth cultures employing the snap-chill method as described by Swetha et al.17 Polymerase Chain Reaction (PCR) confirmation was performed for the commonly observed bacterial contaminants such as S. aureus and Escherichia coli isolates.
Molecular confirmation of S. aureus isolates using PCR
The PCR reaction mixture (12.5 µL) consisted of 6.25 µL of 2X DreamTaq master mix, 8 pmole each of forward and reverse primers, reaction buffer containing 1.5 mM magnesium chloride (MgCl2), 2 µL of DNA template and the volume was made up to 12.5 µL using nuclease-free water. The cyclic thermal conditions targeting the nuc gene for the molecular confirmation of S. aureus were as follows: initial denaturation at 94°C for 5 min followed by denaturation at 94°C for 30 sec, annealing at 57°C for 1 min, extension at 72°C for 1 min for 30 cycles, followed by final extension at 72°C for 7 min. Similarly, thermal conditions targeting the coa gene of S. aureus were as follows: initial denaturation at 94°C for 5 min followed by denaturation at 94°C for 30 sec, annealing at 58°C for 1 min, extension at 72°C for 1 min for 30 cycles, and final extension at 72°C for 7 min.18
Molecular confirmation of E. coli isolates using PCR
The molecular confirmation of E. coli was performed based on PCR targeting cydA (cytochrome bd complex) and uidA (β-D-galactosidase) genes using the conditions as follows: initial denaturation at 94°C for 5 min followed by 30 cycles each of denaturation at 94°C for 30 sec, annealing at 60°C for 30 sec, extension at 72°C for 45 sec and final extension at 72°C for 5 min.19
Determination of antibiotic susceptibility pattern of the isolates
The isolates were subjected to antibiotic susceptibility by the Kirby-Bauer disc diffusion method described by the Clinical and Laboratory Standards Institute (CLSI 2023) (Figure 1).20 The antibiotic discs used for Staphylococcus and Bacillus spp. isolates were gentamicin (10 µg), chloramphenicol (30 µg), enrofloxacin (5 µg), penicillin (10U), erythromycin (15 µg), tetracycline (30 µg), sulphamethoxazole-trimethoprim (1.25/23.75 µg), linezolid (30 µg), cefoxitin (30 µg), and vancomycin (30 µg). The antibiotic discs used for E. coli isolates were ampicillin (10 µg), amikacin (30 µg), ceftazidime (30 µg), cefoxitin (30 µg), cefotaxime (30 µg), ceftriaxone (30 µg), cefpodoxime (10 µg), nalidixic acid (30 µg), enrofloxacin (5 µg), sulphamethoxazole-trimethoprim (1.25-23.75 µg), chloramphenicol (30 µg), and tetracycline (30 µg).
A total of 351 culture flasks belonging to P1-P8 passages during the 2020-2022 period (3 years) were screened for contamination. Among them, only five flasks (1.42%) were found to be contaminated.
Gram’s staining characteristics
On nutrient agar round, creamish-yellow colour colonies were observed, which were subjected to Gram’s staining. Gram-positive cocci, occurring in clusters and Gram-positive rods, occurring in chains were observed. Moreover, Gram-negative rods were also observed from different samples (Table 1).
Table (1):
The results of the gram staining and morphology of the bacteria isolated
No. |
Isolate ID |
Gram character |
Morphology |
|---|---|---|---|
1. |
CCS1 |
Gram-positive |
Cocci arranged in clusters, few in chains also |
2. |
CCS2 |
Gram-positive |
Rods arranged in chains |
3. |
CCS3 |
Gram-negative |
Rods |
4. |
CCS4 |
Gram-positive |
Cocci arranged in short chains |
5. |
CCS5 |
Gram-negative |
Rods |
Colony characteristics on selective media
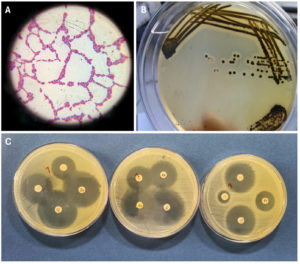
On Baird-Parker Agar (for Staphylococcus spp.), round black colonies with surrounding opaque zones were observed that had a shiny texture (Figure 1). On Eosin Methylene Blue Agar (for E. coli), round green-metallic sheen smooth colonies were observed with a dark centre. On Sheep Blood Agar (for Bacillus spp.) round smooth dull/wrinkled irregular colonies were observed with some colonies showing haemolysis.
Figure 1. (A) Pink cocci in clusters. (B) Black colonies of Staphylococcus spp. on Baird-Parker Agar. (C) Representative picture illustrating phenotypic confirmation of antimicrobial resistance of isolates by disk diffusion method
Biochemical identification of the microorganisms
The results of the biochemical test used for the characterization of the bacterial isolates are given in Table 2. The results are based on the interpretation of positive (+) and negative (-) reactions for each biochemical test, and “NP” indicates that the specific test was not performed.
Table (2):
Result of the biochemical test used for identification of the bacterial isolate. The results are based on the interpretation of positive (+) and negative (-) reactions for each biochemical test, and “NP” indicates that the specific test was not performed
Isolate ID |
Motility |
Catalase |
Oxidase |
Coagulase test |
Indole |
MR |
VP |
Citrate |
TSI |
Urease |
Nitrate reduction |
Presumptive identification |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
CCS1 |
– |
+ |
– |
+ |
– |
+ |
– |
– |
NP |
NP |
NP |
Staphylococcus aureus |
CCS2 |
+ |
+ |
+ |
NP |
– |
– |
+ |
+ |
NP |
– |
+ |
Bacillus spp. |
CCS3 |
+ |
+ |
– |
NP |
+ |
+ |
– |
– |
AG/A acid (yellow) and gas formation in butt and acid (yellow) on slant |
– |
NP |
E. coli |
CCS4 |
– |
+ |
– |
+ |
– |
+ |
– |
– |
NP |
NP |
NP |
Staphylococcus aureus |
CCS5 |
+ |
+ |
– |
NP |
+ |
+ |
– |
– |
AG/A acid (yellow) and gas formation in butt and acid (yellow) on slant |
– |
NP |
E. coli |
Molecular identification of the bacterial isolates
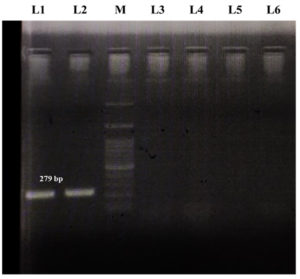
An amplicon size of 279 bp was observed in the S. aureus isolates based on the nuc gene and an amplicon size of 600 bp was observed based on the coa gene. Similarly, an amplicon size of 398 bp and 603 bp was observed in the E. coli isolates for cydA and uidA genes, respectively. A representative image of the agarose gel electrophoresis of the PCR amplicon of the nuc gene of S. aureus is depicted in Figure 2.
Figure 2. PCR confirmation of S. aureus isolates based on nuc gene. Lane L1 and L2: Positive amplicon for nuc gene, Lane M: 100 bp ladder, and Lane L3 to L6: Blank
Antibiotic susceptibility pattern
The antibiotic susceptibility patterns of different isolates are given in Tables 3 and 4. For isolate CCS1, susceptibility was observed to gentamicin, chloramphenicol, ciprofloxacin, tetracycline, trimethoprim-sulfamethoxazole, linezolid, cefoxitin, and vancomycin. CCS2 displayed susceptibility to gentamicin, chloramphenicol, streptomycin, tetracycline, trimethoprim-sulfamethoxazole, linezolid, and vancomycin. Isolate CCS4 exhibited susceptibility to gentamicin, streptomycin, tetracycline, linezolid, cefoxitin, and vancomycin. Moving to Table 4, isolates CCS3 demonstrated susceptibility to ceftazidime, nalidixic acid, trimethoprim-sulfamethoxazole, and tetracycline. Lastly, CCS5 displayed susceptibility to amoxicillin-clavulanate, ceftazidime, nalidixic acid, ciprofloxacin, tetracycline, trimethoprim-sulfamethoxazole, and tetracycline. These results provide crucial information regarding the antibiotic susceptibility profiles of the bacterial contaminants, aiding in the development of effective treatment strategies and the maintenance of aseptic conditions in SCCs.
Table (3):
The antibiotic susceptibility pattern of different isolates (Staphylococcus and Bacillus spp. isolates)
Isolate ID |
GEN |
C |
E |
P |
EX |
TE |
SXT |
LZ |
CX |
VA |
|---|---|---|---|---|---|---|---|---|---|---|
CCS1 |
S |
S |
R |
I |
S |
S |
S |
S |
S |
S |
CCS2 |
S |
S |
R |
R |
S |
S |
S |
S |
R |
S |
CCS4 |
S |
I |
I |
R |
S |
S |
R |
S |
S |
S |
Table (4):
The antibiotic susceptibility pattern of different isolates (E. coli isolates)
Isolate ID |
AMP |
AM |
CAZ |
CX |
CTX |
CTR |
CPD |
NA |
EX |
SXT |
C |
TE |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
CCS3 |
R |
R |
S |
I |
R |
I |
R |
S |
I |
S |
R |
S |
CCS5 |
I |
S |
S |
R |
R |
R |
I |
S |
S |
S |
S |
S |
Quality assurance in stem cell laboratories involves comprehensive testing protocols to assess cell viability, identity, potency, and purity, ensuring that the final product meets predefined standards.15 From the initial stages of cell collection to their manipulation, expansion, and eventual therapeutic application, every step follows established standard operating procedures (SOPs). These SOPs are continuously refined based on scientific advancements and regulatory guidelines.21 Through multifaceted measures, quality assurance in stem cell labs not only aims for consistent high-quality outcomes but also prioritizes patient safety and therapeutic effectiveness.
Regular checks for contaminants are imperative in stem cell laboratories due to the sensitivity and vulnerability of cell cultures to external agents.22,23 These checks are mandated to uphold the integrity and safety of cell-based products. SCCs, often derived from biological sources like bone marrow or adipose tissue, are highly susceptible to microbial contamination by bacteria, fungi, or other microorganisms.24 Even minute levels of contaminants can alter cell behaviour, compromise experimental outcomes, or pose health risks if introduced into therapeutic applications. Therefore, routine monitoring through microbial surveillance helps to detect, identify, and mitigate any potential contaminants promptly.25 By conducting these checks regularly, laboratories ensure the maintenance of sterile environments, validate the quality of cell cultures and uphold the reliability and safety standards required for both research and clinical applications involving stem cells.26,27
Isolating and identifying bacterial contaminants in SCCs involves meticulous steps to ensure the purity and integrity of these delicate cultures.22 Initially, contamination suspicion prompts the collection of samples using sterile tools like swabs or pipettes from the affected area of the culture. These samples are then inoculated onto agar plates containing specific growth media suitable for bacterial growth. Incubation follows, allowing bacterial colonies to flourish, which are subsequently observed and isolated for purity through subculturing techniques.13,24 Identification involves various methods, including biochemical assays, molecular techniques like PCR, or utilizing diagnostic kits to pinpoint the specific bacterial strain. This comprehensive process demands precision, adherence to sterile conditions, and expertise in microbiological methods to accurately isolate and identify contaminants, ensuring the preservation of the pristine nature of the SCC.27 Utilizing PCR primers for bacterial contaminant detection significantly bolsters the quality control measures of SCCs, reducing the risk of inadvertent contamination spread to healthy cultures during tissue culture processes.28
Common bacterial contaminants in SCCs encompass various species that can compromise the integrity and safety of these delicate cultures.22 Our study indicated that the common bacterial contaminants of R-BMSC cultures include, species like S. aureus, Bacillus spp. and E. coli. These bacteria can inadvertently infiltrate cultures during cell isolation, handling, or processing stages. S. aureus, a common skin bacterium, might enter cultures through improper aseptic techniques during isolation.29 E. coli, predominantly found in the gastrointestinal tract, could contaminate cultures due to inadequate sterilization of instruments or reagents.12 Bacillus spp. are ubiquitous inhabitants of the surroundings, and their presence in culture flasks can be detrimental to the stem cells. Contamination can occur during culture handling, media preparation, or from laboratory surfaces.30 Bacillus spp. is particularly resilient and can form heat-resistant spores, making them challenging to eradicate through routine sterilization methods. Their introduction into SCCs may lead to alterations in cell behaviour, affecting the reliability and reproducibility of research outcomes.31
The determination of antibiotic susceptibility patterns of the contaminating isolates, specifically Staphylococcus and Bacillus spp., becomes crucial in understanding their resistance profiles and devising appropriate strategies to eliminate or control these contaminants.12 Similarly, understanding the antibiotic susceptibility pattern of E. coli isolates is essential given their potential presence and impact on SCCs.32 The selection of a wide range of antibiotics in this study, including gentamicin, chloramphenicol, enrofloxacin, penicillin, erythromycin, tetracycline, sulphamethoxazole-trimethoprim, linezolid, cefoxitin, vancomycin, ampicillin, amikacin, ceftazidime, cefotaxime, ceftriaxone, cefpodoxime, nalidixic acid, and enrofloxacin, allows for comprehensive assessment of antibiotic susceptibility in the isolated strains.33 Interpreting the susceptibility patterns observed in these isolates will aid in determining the most appropriate antibiotic(s) for treatment or eradication strategies within SCCs. It is crucial to identify antibiotics to which these isolates are susceptible, ensuring the preservation of the integrity of SCCs without compromising the health or functions of cells.34
The study aimed to isolate the contaminants in the SCC derived from rabbit bone marrow. We isolated and identified Bacillus spp. and E. coli as the common contaminants based on isolation, phenotypic and molecular characterization. We have changed the antimicrobial protocol based on the results of the antibiotic susceptibility test and monitored further for bacterial contamination and viability of SCCs. It is worth mentioning that once the changes were implicated in adherence to SOPs, further no contaminations were detected upon routine monitoring for the past two years.
This research advances our understanding of bacterial contaminants in MSC cultures, offering valuable insights for maintaining the integrity of these cultures in regenerative medicine and cell-based therapies. The isolation and characterization of bacterial contaminants presented herein contribute to the establishment of stringent quality control measures necessary for the successful translation of MSC research into clinical applications. The determination of antibiotic susceptibility patterns of contaminating isolates in SCCs, especially S. aureus, Bacillus spp., and E. coli, is a critical step in ensuring the reliability and validity of experimental results. Rigorous monitoring and stringent aseptic techniques are essential in R-BMSCs to prevent common bacterial contaminants from compromising the quality and safety of the cell cultures. Although the identification technique is straightforward but is time-consuming. Except this, the procedure exhibits exceptional efficacy, ensuring a 100% success rate in eliminating bacteria from treated SCCs. Further investigations into preventive strategies and novel detection methods will be crucial for ensuring the safety and efficacy of MSC-based therapies.
ACKNOWLEDGMENTS
The authors would like to thank Director, ICAR-Indian Veterinary Research Institute, Izatnagar, Bareilly, India, for providing the necessary research facilities to carry out this work.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This study was carried out according to the guidelines approved by the Institute Animal Ethics Committee (IAEC), Indian Veterinary Research Institute (ICAR-IVRI), Izatnagar, Bareilly, Uttar Pradesh, India.
- Musial-Wysocka A, Kot M, Majka M. The Pros and Cons of Mesenchymal Stem Cell-Based Therapies. Cell Transplant. 2019;28(7):801-812.
Crossref - Sharun K, Musa TH, Musa HH, et al. Mapping global trends in adipose-derived mesenchymal stem cell research: A bibliometric analysis using scopus database. Ann Med Surg (Lond). 2022;77:103542.
Crossref - Banu SA, Pawde AM, Sharun K, et al. Evaluation of bone marrow-derived mesenchymal stem cells with eggshell membrane for full-thickness wound healing in a rabbit model. Cell Tissue Bank. 2023.
Crossref - Sivanarayanan TB, Bhat IA, Sharun K, et al. Allogenic bone marrow-derived mesenchymal stem cells and its conditioned media for repairing acute and sub-acute peripheral nerve injuries in a rabbit model. Tissue Cell. 2023;82:102053.
Crossref - Sharun K, Rawat T, Kumar R, et al. Clinical evaluation following the percutaneous transplantation of allogenic bone marrow-derived mesenchymal stem cells (aBM-MSC) in dogs affected by vertebral compression fracture. Vet Anim Sci. 2020;10:100152.
Crossref - Bist D, Pawde AM, Amarpal, et al. Evaluation of canine bone marrow-derived mesenchymal stem cells for experimental full-thickness cutaneous wounds in a diabetic rat model. Expert Opin Biol Ther. 2021;21(12):1655-1664.
Crossref - Peer BA, Bhat AR, Shabir U, et al. Comparative evaluation of fracture healing potential of differentiated and undifferentiated guinea pig and canine bone marrow-derived mesenchymal stem cells in a guinea pig model. Tissue Cell. 2022;76:101768.
Crossref - Sharun K, Muthu S, Mankuzhy PD, et al. Cell-free therapy for canine osteoarthritis: current evidence and prospects. Vet Q. 2022;42(1):224-230.
Crossref - Yuan BZ. Establishing a Quality Control System for Stem Cell-Based Medicinal Products in China. Tissue Eng Part A. 2015;21(23-24):2783-2790.
Crossref - Kallur T, Blomberg P, Stenfelt S, Tryggvason K, Hovatta O. Quality Assurance in Stem Cell Banking: Emphasis on Embryonic and Induced Pluripotent Stem Cell Banking. Methods Mol Biol. 2017;1590:11-16.
Crossref - Inamdar MS, Healy L, Sinha A, Stacey G. Global solutions to the challenges of setting up and managing a stem cell laboratory. Stem Cell Rev Rep. 2012;8(3):830-843.
Crossref - Cobo F, Stacey GN, Hunt C, et al. Microbiological control in stem cell banks: approaches to standardisation. Appl Microbiol Biotechnol. 2005;68(4):456-466.
Crossref - Meechan PJ, Potts J. Biosafety in microbiological and biomedical laboratories, 6th edn. US Department of Health and Human Services. Public Health Service, Centers for Disease Control and Prevention, National Institutes of Health, Atlanta, GA. www. cdc. gov/labs/BMBL. html. 2020.
- Geraghty RJ, Capes-Davis A, Davis JM, et al. Guidelines for the use of cell lines in biomedical research. Br J Cancer. 2014;111(6):1021-1046.
Crossref - Volarevic V, Markovic BS, Gazdic M, et al. Ethical and Safety Issues of Stem Cell-Based Therapy. Int J Med Sci. 2018;15(1):36-45.
Crossref - Parry JM, Turnbull PC, Gibson JR. A colour atlas of Bacillus species. Wolfe Medical Publications Ltd. 1983.
- Swetha CS, Rao TM, Babu AJ, Kumar E. Process Optimization for the Detection of Listeria monocytogenes and Listeriolysin O from Spiked Chicken Meat by Polymerase Chain Reaction. J Meat Sci. 2015;10(2):7-15.
- Javid F, Taku A, Bhat MA, Badroo GA, Mudasir M, Sofi TA. Molecular typing of Staphylococcus aureus based on coagulase gene. Vet World. 2018;11(4):423-430.
Crossref - Horakova K, Mlejnkova H, Mlejnek P. Specific detection of Escherichia coli isolated from water samples using polymerase chain reaction targeting four genes: cytochrome bd complex, lactose permease, beta-D-glucuronidase, and beta-D-galactosidase. J Appl Microbiol. 2008;105(4):970-976.
Crossref - Tamma PD, Harris PN, Mathers AJ, Wenzler E, Humphries RM. Breaking down the breakpoints: rationale for the 2022 Clinical and Laboratory Standards Institute revised piperacillin-tazobactam breakpoints against Enterobacterales. Clin Infect Dis. 2023;77(11):1585-1590.
Crossref - Wesselschmidt RL, Schwartz PH. The stem cell laboratory: design, equipment, and oversight. Methods Mol Biol. 2011;767:3-13.
Crossref - Cobo F, Cortes JL, Cabrera C, Nieto A, Concha A. Microbiological contamination in stem cell cultures. Cell Biol Int. 2007;31(9):991-995.
Crossref - Senthil V, Paul A. Stem Cell Research-A Review on the Latest Indian Guidelines. J Pharm Sci Res. 2019;11(4):1160-1164.
- Nikfarjam L, Farzaneh P. Prevention and detection of Mycoplasma contamination in cell culture. Cell J. 2012;13(4):203-212.
- Sekiya EJ, Forte A, Kühn TI, Janz F, Bydlowski SP, Alves A. Establishing a stem cell culture laboratory for clinical trials. Rev Bras Hematol Hemoter. 2012;34(3):236-241.
Crossref - Izarra ML, Panta AL, Maza CR, et al. Identification and control of latent bacteria in in vitro cultures of sweetpotato [Ipomoea batatas (L.) Lam]. Front Plant Sci. 2020:903.
Crossref - Franco-Duarte R, Cernakova L, Kadam S, et al. Advances in Chemical and Biological Methods to Identify Microorganisms-From Past to Present. Microorganisms. 2019;7(5):130.
Crossref - Tokuno O, Hayakawa A, Yanai T, et al. Sterility Testing of Stem Cell Products by Broad-Range Bacterial 16S Ribosomal DNA Polymerase Chain Reaction. Lab Med. 2015;46(1):34-41.
Crossref - Willis ZI, Brondon JE, Sickbert-Bennett EE, Kasow KA, Weber DJ. Staphylococcus aureus Bloodstream Infection Due to Contaminated Hematopoietic Stem-Cell Graft. Infect Control Hosp Epidemiol. 2018;39(3):367-369.
Crossref - Nims RW, Price PJ. Best practices for detecting and mitigating the risk of cell culture contaminants. In Vitro Cell Dev Biol Anim. 2017;53(10):872-9.
Crossref - Mahmood A, Ali S. Microbial and viral contamination of animal and stem cell cultures: common contaminants, detection and elimination. J Stem Cell Res Ther. 2017;2(5):1-8.
Crossref - Marrazzo P, Pizzuti V, Zia S, et al. Microfluidic Tools for Enhanced Characterization of Therapeutic Stem Cells and Prediction of Their Potential Antimicrobial Secretome. Antibiotics (Basel). 2021;10(7):750.
Crossref - Franca L, Simoes C, Taborda M, Diogo C, da Costa MS. Microbial Contaminants of Cord Blood Units Identified by 16S rRNA Sequencing and by API Test System, and Antibiotic Sensitivity Profiling. PLoS One. 2015;10(10):e0141152.
Crossref - Pamies D, Bal-Price A, Simeonov A, et al. Good Cell Culture Practice for stem cells and stem-cell-derived models. ALTEX. 2017;34(1):95-132.
Crossref
© The Author(s) 2024. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.