ISSN: 0973-7510
E-ISSN: 2581-690X
Fourteen potential antagonist bacterial isolates were obtained from soil samples collected from the rhizospheric soil of healthy chickpea. Colony and morphological characterization revealed that all isolates were diversified and distinct from each other. The antagonism characters against Fusarium wilt pathogen of theses isolates were very prominent and ranged from 21.7 to 75.0%, seven out of fourteen isolates were showing more than 50.0% inhibition of Fusarium oxysporum under In vitro assay. These seven highly potential bacterial isolates were identified by 16s rRNA sequencing out of which three highly effective antagonistic isolates were identified as Burkholderia sp. (K10), Bacillus subtilis (K18) and Bacillus aryabhattai (K21) showing 71.2, 75.0 and 70.2% antagonism, respectively. The plant growth promoting characters of all the isolates viz., nitrogen fixation, phosphate solubilization, zinc solubilization, siderphore production and IAA production were studied. Highly effective antagonist isolates K18 and K21 were also efficient for N2 fixation and Zn solubilization. These two promising antagonist isolates with PGPR activities could be further evaluated in field study for their commercial exploitation.
Antagonist, biocontrol, Fusarium oxysporum f. sp. ciceris.
Rhizosphere region harbours bewildering diversity of different group of microorganisms. It is a potential place for complex plant-microbe and microbe-microbe interactions1. Thus, rhizosphere region is an extremely complex habitat for microbes known as Plant Growth Promoting Rhizobacteria (PGPR), plant beneficial bacteria. Many PGPR helps plant to impart resistance to biotic stresses by antagonism mechanism i.e. interference with plant pathogen growth, survival, infection or plant attack.2 Antagonistic bacteria can inhibit plant pathogens by different mechanisms like inhibition of the pathogen by antibiotics and toxins, competition for space and nutrients, parasitism by extracellular cell-wall-degrading enzymes, mycophagy, etc.1,3
Chickpea (Cicer arietinum L.) is pivotal source of protein for vegetarian diet, however the productivity of chickpea is far below its potential.4 One of the major reasons for the low productivity of cultivated chickpea is its narrow genetic bases and its sexual incompatibility with other wild species of Cicer in natural inter specific cross. Furthermore, various biotic (Fusarium wilt, Aschochyta blight, nematodes and pests) and abiotic (heat, salinity, drought and cold) stresses severely reduce the yield5. Among the biotic stresses, the Fusarium wilt and Ascochyta blight are the most important fungal diseases causing serious yield losses. Fusarium wilt caused by Fusaruim oxysporum f. sp. ciceris (Padwick) Matuo & K. Sato (FOC) is one of the most important and destructive vascular disease of chickpea. Fusarium wilt is primarily managed by resistance breeding programs but pathogenic variability and mutability leading to breakdown of naturally selected resistance, are the main hurdles for plant breeders.6 The fungicides are used for wilt management however, the degradation of fungicides and organic compounds are very difficult and it’s accumulation in food chains had higher toxic level in animals and also lead to environment pollution.7 Therefore, Integrated Disease Management (IDM) strategy is need of time which includes minimum use of chemicals for checking the pathogen population, encouragement of beneficial biological agents to reduce the pathogen inoculums8, further the crop rotation, pathogen-free seed, removal of plant debris and fungicide seed treatment are several disease management strategies have been employed for the control of wilt in chickpea. Biological control of Fusarium wilt is a potential component of IDM where the principle of bacteria or fungal antagonist has been exploited for the control of disease. The application of microorganisms to control diseases, which is a form of biological control, is an environment friendly approach. Realizing the need of novel microbial biocontrol agent against the Fusarium wilt of chickpea, the present study is undertaken with an objective to isolate efficient biocontrol agent with PGPR activity from chickpea rhizosphere.
Collection of samples
Different soil samples were collected from the rhizosphere region of healthy chickpea plant from pulse research station, Navsari Agricultural University, Navsari 396 450, Gujarat, India.
Isolation of PGPR
Collected soil samples were serially diluted, plated on N-agar medium and incubated for 24 hours. After incubation, well isolated and distinct colonies were streaked on N-agar plates by four sector method9. Antagonist effect of all isolates was studied against F. oxysporum following the method described by Jinantana and Sariah1997.10 Based on initial screening for antagonism against Fusarium oxysporum f. sp. ciceris, total fourteen distinct isolates showing considerable antagonism were preserved on agar slant at 4°C and glycerol stocks were prepared for long term preservation of isolates.
Characterization of isolates
All the isolates were characterized in terms of colony and morphological characters to judge diversity and distinctness among isolates.
In vitro screening of isolates
Isolates were screened for their antagonist as well as plant growth promoting activities such as nitrogen fixation, phosphate and zinc solubilization, siderophore production and IAA production.
Antagonistic activity
Antagonistic activities of the isolates were evaluated by dual culture method against Fusarium oxysporum f. sp. ciceris. Fungal and bacterial isolates were inoculated on Potato Dextrose Agar (PDA) medium in test petri dish and inoculation of only fungal pathogen on PDA served as a control. Plates were incubated at 30°C temperatures till full coverage of fungi in control. Efficiency of isolates was judged on per cent growth inhibition method given by Vincent, 1947.11
PGI=C-T/C X 100
Where,
PGI= Percent Growth Inhibition
T= Colony diameter of pathogenic fungi in test plate (mm)
C= Colony diameter of pathogenic fungi in Control plate (mm)
Nitrogen fixation
Nitrogen fixation ability of isolates was judged by inoculating pure culture on nitrogen free Ashby’s Mannitol Agar medium. Presence of growth after 48 hrs of incubation at 28°C temperature was taken as evidence of positive nitrogen fixation reaction.
Phosphate solubilization
Rate of phosphate solubilization was checked by inoculating bacteria on Pikovskya’s medium by spot test method followed by measurement of zone ratio.12 Medium possesses insoluble tricalcium phosphate, which on acidification, gives clear zone around colony.
Zinc solubilization
Zinc solubilization efficacy was judged on modified Pikovskaya medium.13 Pure culture was applied by spot test method and efficacy was judged on the basis of zone of clearance of insoluble zinc oxide substrate.
Siderophore production
Siderophore production was judged by spot inoculation of 1µl bacterial suspension on chrome azurole S agar plates and incubated at 30°C temperature for 5 days in dark condition. Appearance of orange halogen around the colony was considered positive test for siderophore production.14
IAA production
IAA estimation was done using Salkowaskay method.15 A loopful of culture was inoculated in 2 ml Minimal Media amended with 50 µg/ml tryptophan. Inoculated broths were incubated for 24 hrs at 28°C temperature on rotary shaker, centrifuged at 10,000g for 15 minutes, 1ml of supernatant was taken in fresh tube and 2 to 3 drops of Ortho-phosphoric acid was added, 2ml of Salkowaskay reagent (1ml of 0.5M FeCl3 in 50 ml of 35% HClO4) was added to the aliquot. Samples were incubated for 25 minutes at room temperature. Absorbance was measured at 530 nm. Auxin quantification values were recorded by preparing calibration curve made by using IAA standard in the range of 10-100 µg/ml.
Identification of Selected Effective Microbes’ Bacteria
Total bacterial genomic DNA was extracted and purified according to the CTAB method [16] with slight modifications. The 16S rRNA genes were amplified from genomic DNA with the primers 27F: 5-AGAGTTTGATCCTGGCTCAG-3 and 1492R: 5-CGGTTACCTTGTTACGACTT-3. PCR amplification was performed as follows: initial denaturation 5 min at 94°C, followed by 32 cycles of 94°C for 30s, 30s at 40°C, 90s at 72°C, and final extension for 5 min at 72°C, at the last hold at 4°C. Sequencing of amplified 16S rRNA was done using AB prism Sequencer, 3130 Genetic analyzer (Applied Bio systems) with 4 capillaries. DNA sequence homology searches were performed using the online BLAST search engine in GenBank (available at: http://www.ncbi.nlm.nih.gov). Phylogenetic analyses were conducted by using phylogeny.fr17
Antagonistic microbes have the potential to inhibit plant pathogenic microorganisms by different mechanisms in eco-friendly manner. Isolation of such important microorganisms was performed from soil samples collected from healthy chickpea rhizospheric soil. Total fourteen rhizospheric bacteria with distinct colony and morphological characteristics were obtained by dilution plate techniques (Table 1) and preserved for future studies.
Table (1):
Colony and morphological characters of potential rhizospheric isolates.
| Sr. no. | Culture ID | Colony characters | Morphological characters | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Size | Elevation | Shape | Margin | Opacity | Consistency | Pigmentation | Texture | Grams reaction | Shape | Arrangement | ||
| 1 | K 1 | Medium | Raised | Round | Entire | Translucent | Sticky | White | Smooth | Negative | Short Rod | Single |
| 2 | K 2 | Medium | Raised | Round | Entire | Translucent | Sticky | Red | Smooth | Positive | Cocci | Single |
| 3 | K 8 | Medium | Raised | Round | Entire | Translucent | Sticky | White | Smooth | Positive | Long Rod | Pair |
| 4 | K 10 | Big | Slightly Raised | Irregular | Entire | Transparent | Sticky | Colourless | Smooth | Negative | Short Rod | Single |
| 5 | K 11 | Medium | Flat | Oblong | Entire | Translucent | Rough | White | Rough | Positive | Long Rod | Chain |
| 6 | K 12 | Medium | Flat | Irregular | Undulate | Translucent | Sticky | White | Smooth | Positive | Long Rod | Single |
| 7 | K 14 | Medium | Raised | Round | Entire | Transparent | Sticky | Colourless | Smooth | Negative | Short Rod | Single |
| 8 | K 15 | Small | Raised | Irregular | Irregular | Translucent | Rough | White | Rough | Positive | Long Rod | Single |
| 9 | K 17 | Small | Raised | Irregular | Irregular | Translucent | Rough | White | Rough | Positive | Long Rod | Chain |
| 10 | K 18 | Big | Flat | Irregular | Entire | Translucent | Rough | White | Rough | Positive | Long Rod | Pair |
| 11 | K 19 | Medium | Raised | Round | Entire | Translucent | Sticky | Pinkish | Smooth | Positive | Long Rod | Pair |
| 12 | K 20 | Small | Slightly Raised | Irregular | Entire | Translucent | Sticky | White | Rough | Positive | Long Rod | Pair |
| 13 | K 21 | Small | Raised | Round | Irregular | Translucent | Sticky | White | Sticky | Positive | Long Rod | Single |
| 14 | K 22 | Big | Flat | Round | Entire | Translucent | Rough | White | Rough | Positive | Long Rod | Single |
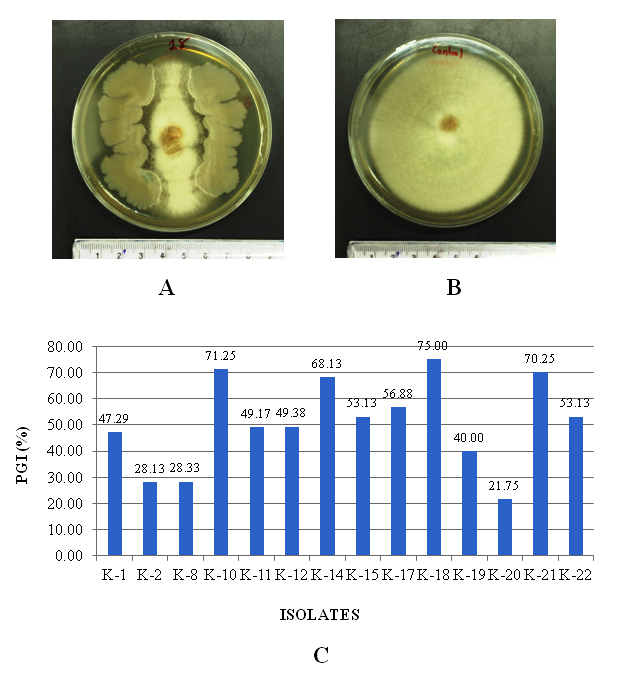
Use of fungicides in the plant disease management is not advantageous because of its ill effect on environment and non-targeted organisms.18 Use of antagonistic microbes in plant disease management can be the best alternate of fungicides with eco-friendliness. Antagonistic microbes were screened by dual culture method on PDA agar medium against Fusarium oxysporum causing wilt in chickpea, the per cent antagonist range from 21.75 to 75.00%. out of which three isolates viz. K18, K10 and K21 showed higher biocontrol potential which was 75.00, 71.25 and 70.25 per cent, respectively, while the lowest efficacy was reported in isolate K20 (21.75%; Fig. 1).
Fig. 1. A. Plate of K18 isolate dual culture after 7 days incubation; B. Control plate of F. oxysporum after 7 days incubation; C. Antagonistic potential of different rhizospheric isolates against Fusarium oxysporum f. sp.
For the characterization of plant growth promotion characters, isolates were also checked for the efficacy to increase the availability of different nutrients i.e. nitrogen, phosphorus and zinc (Table 2). The isolate K2 and K17 were found positive for increasing the availability of N2, PO4 and Zn, the isolate K12 and K20 were found positive in nitrogen fixation only, while remaining isolates K1, K8, K11, K14, K15, K18 and K21 were able to increase the availability of N2 and Zn. Moreover isolated K19 and K22 were also found promising for phosphate solubilization in lab medium with potential antagonistic activity against Fusarium wilt pathogen of chickpea. Iron is a vital element required by every living organisms as a part of electron transport chain and as a cofactor of different enzymes.19 Siderohpore producing bacteria produces compound that can chelate the iron element and thus make it unavailable for plant pathogens in rhizospheric region. Siderophore producing bacteria thus works as a biocontrol agent for plant pathogens. Among the different isolates, the isolate viz. K2, K10, K15 and K17 were found to be siderophore positive. Indole 3 Acetic Acid (IAA) is a plant growth promoter and produced by many rhizospheric bacteria, however, none of 14 antagonistic bacteria were found to produce IAA in lab medium amended with tryptophan (Table 3).
Table (2):
Efficacy of different isolates for increasing availability of nutrients.
Sr. no. |
Culture ID |
Nitrogen fixation |
Phosphate solubilization |
Zinc solubilization |
|---|---|---|---|---|
1 |
K 1 |
Positive |
Negative |
Positive |
2 |
K 2 |
Positive |
Positive |
Positive |
3 |
K 8 |
Positive |
Negative |
Positive |
4 |
K 10 |
Negative |
Negative |
Negative |
5 |
K 11 |
Positive |
Negative |
Positive |
6 |
K 12 |
Positive |
Negative |
Negative |
7 |
K 14 |
Positive |
Negative |
Positive |
8 |
K 15 |
Positive |
Negative |
Positive |
9 |
K 17 |
Positive |
Positive |
Positive |
10 |
K 18 |
Positive |
Negative |
Positive |
11 |
K 19 |
Negative |
Positive |
Negative |
12 |
K 20 |
Positive |
Negative |
Negative |
13 |
K 21 |
Positive |
Negative |
Positive |
14 |
K 22 |
Negative |
Positive |
Negative |
Table (3):
Efficacy of different isolates for Siderophore and IAA production.
Sr. no. |
Culture ID |
Siderphore production |
IAA production |
|---|---|---|---|
1 |
K 1 |
Negative |
Negative |
2 |
K 2 |
Positive |
Negative |
3 |
K 8 |
Negative |
Negative |
4 |
K 10 |
Positive |
Negative |
5 |
K 11 |
Negative |
Negative |
6 |
K 12 |
Negative |
Negative |
7 |
K 14 |
Negative |
Negative |
8 |
K 15 |
Positive |
Negative |
9 |
K 17 |
Positive |
Negative |
10 |
K 18 |
Negative |
Negative |
11 |
K 19 |
Negative |
Negative |
12 |
K 20 |
Negative |
Negative |
13 |
K 21 |
Negative |
Negative |
14 |
K 22 |
Negative |
Negative |
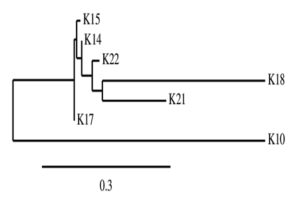
The 16S rRNA sequencing of seven potential antagonist bacterial isolates was done for identification. The PCR products of approximately 1380 and 1270 bp were generated for the seven effective antagonistic bacteria. The partial 16S rDNA sequences of all isolates were determined and aligned to other known sequences database in GenBank for their identification (Table 4). The three effective anatagonist K10, K18 and K21 were identified as Burkholderia sp., Bacillus subtilis and Bacillus aryabhattai, respectively. The phylogenetic analysis of all seven antagonistic microorganisms using 16s rRNA sequence revealed maximum similarity between isolate K18 and K21 (Figure 2).
Fig. 2. Phylogenic relationship among seven highly efficient antagonistic isolated based on 16s rRNA sequence
Table (4):
Identification of potential antagonistic microorganism using 16S rRNA sequence.
Culture ID |
Organism identified by BLAST |
Query Coverage (%) |
E Value |
Accession no. |
|---|---|---|---|---|
K10 |
Burkholderia sp. SBH-11 gene for 16S ribosomal RNA, partial sequence |
74 |
6e-50 |
AB366333.1 |
K14 |
Bacillus subtilis strain MML2483 16S ribosomal RNA gene, partial sequence |
98 |
0.0 |
KJ655546.1 |
K15 |
Bacillus subtilis strain MML2483 16S ribosomal RNA gene, partial sequence |
97 |
0.0 |
KJ655546.1 |
K17 |
Bacillus subtilis strain OTPB4 16S ribosomal RNA gene, partial sequence |
97 |
0.0 |
KT265082.1 |
K18 |
Bacillus subtilis strain I55 16S ribosomal RNA gene, partial sequence |
64 |
2e-39 |
KF318812.1 |
K21 |
Bacillus aryabhattai strain JPR39 16S ribosomal RNA gene, partial sequence |
97 |
0.0 |
KR045599.1 |
K22 |
Bacillus sp. RA23 16S ribosomal RNA gene, partial sequence |
98 |
0.0 |
FJ898306.1 |
The antagonistic effects of twelve rhizosphere isolates (six Bacillus subtilis and six Pseudomonas spp.) against Fusarium oxysporum f. sp. ciceris were studied under in vitro condition, and found P. aeuroginosa (P10 and P12), B. subtilis (B1, B6, B28 and B99) and P. aeuroginosa (P12 and B28) as a potential biocontrol agentL.20 The suppression of chickpea Fusarium wilt by Bacillus subtillis and Trichoderma harzianum treatments in liquid and seed inoculation methods was reported.21 The antagonistic effect of eight antagonistic microorganisms’ viz., Aspergillus flavus, Aspergilus niger, Aspergilus ochraceus, Azotobacter sp., Penicillium sp., Pseudomonas fluorescens, Rhizobium sp. and Trichoderma harzianum was determined under in vitro condition against Fusarium oxysporium f. sp. ciceris.22 All the antagonists reduced the growth of Fusarium oxysporum f. sp. ciceris significantly but Trichoderma harzianum produced longer inhibition zone (6.72 cm) as compared to other antagonistic organisms. The effects of different antagonists, i.e. Acrophialophora sp., fluorescent pseudomonas isolates 1, 3, 4 and Gliocladium virens, G. catanaltum, G. deliguences, Trichoderma hamatam, T. harziamum and T. koningii was screened against the chickpea wilt pathogen, and found that T. hamatam inhibited an average of 26.03 per cent of radial growth, followed by fluorescent pseudomonas-3 (25.5%) and fluorescent pseudomonas isolate-4 (25.08%), respectively.23 The antagonistic activity of 21 isolates of Rhizobium under both in vitro in dual culture and in vivo in green house and field conditions was studied against Fusarium oxysporum f. sp. ciceris race 0. In dual culture study reported that 14 isolates inhibited the mycelia growth of pathogen more than 30% and most effective isolated were Rh8, Rh11, Rh16 and PchSOM, which inhibited fungal growth more than 50 per cent.24 In present study, fourteen isolates were finalized based on antagonistic efficacy and distinguishes, of which only three isolates were found to be potential biocontrol agent under in vitro study, they also showed potential PGPR activities.
Antagonistic microorganisms can be useful for the eco-friendly plant disease management in sustainable agriculture. Apart from antagonistic activity many biocontrol agents also posses PGPR activities, which exert plant growth promotion by direct or indirect mechanisms. In present laboratory study, isolates K17, K18, K21 and K22 obtained from different rhizospheric soil samples were potent in terms of biocontrol and plant growth promotion potential. A combination of these isolates may have biocontrol as well as growth promotion activities like N2 fixation, Zn solubilization, PO4 solubilization and siderophore production. However, potential isolates need to be characterized in field condition for mass exploration.
- Whipps, J. Microbial interactions and biocontrol in the rhizosphere. J. Expr. Bot., 2001; 52: 487–511.
- Chernin, L., Chet, I. Microbial enzymes in biocontrol of plant pathogens and pests. Enzymes in the Environment: Activity, Ecology, and Applications (BurnsR & DickR, eds), Marcel Dekker, Inc., New York. 2002; pp 171–225.
- De Boer, W., Folman, L.B., Summerbell, R.C., Boddy, L. Living in a fungal world: impact of fungi on soil bacterial niche development. FEMS Microbiol. Rev., 2005; 29: 795–811.
- Abbo, S., Berger, J., Turner, N.C. Evolution of cultivated chickpea: four bottlenecks limit diversity and constrain adaptation. Funct. Plant. Biol. 2003; 30: 1081-1087.
- Winter, P., Benko-Iseppon, A.M., Huttel, B., Ratnaparkhe, M., Tullu, A., Sonnante, G., Tekeoglu, M., Santra, D., Sant, V.J., Rajesh, P. N., Kahl, G., Muehlbauer, F.J. A linkage map of the chickpea (Cicer arietinum L.) genome based on 44 recombinant inbred lines from C. arietinum x C. reticulatm cross: localization of resistance genes for Fusarium wilt races 4 and 5. Theoret. Appl. Genet., 2000; 101: 1155–1163.
- Nimbalkara, S.B., Harsulkara, A.M., Giria, P.A., Sainania, M.N., Franceschib, V., Gupta, V.S. Differentially expressed gene transcripts in roots of resistant and susceptible chickpea plant (Cicer arietinum L.) upon Fusarium oxysporum infection. Physiol. Mol. Plant Path., 2006; 68(4-6): 176-188.
- Chet, I. Trichoderma application, mode of action and potential as a biocontrol agent of soil-borne plant pathogenic fungi. In: Innovative approaches to plant disease control, ed. Chet I., Wiley, New York, 1987; pp 137-160.
- Landa, B.B., Navas-Cortes, J.A., Jimenez-Diaz, R.M. Integrated management of Fusarium wilt of chickpea with sowing date, host resistance, and biological control. Phytopathology, 2004; 94: 946-960.
- Montealegre, J.R., Reyes, R., Perez, L.M., Herrera, R., Silva, P., Besoain, X. Selection of bioantagonistic bacteria to be used in biological control of Rhizoctonia solani in tomato. Electron. J. Biotechnol., 2003; 6: 116-127.
- Jinantana, J., Sariah M. Antagonistic effect of Malaysian isolates of Trichoderma harzianum and Gliocladiwn virens on Sclerotium rolfsii”. Pertanika J. Trop. Agric. Sci., 1997; 20: 35-41.
- Vincent, J.M. The esters of 4-hydroxybenzoic acid and related compounds. Part I. Methods for the study of their fungistatic properties, J. Soc. Chem. Ind., 1947; 66: 149-155.
- Pikovskaya, R.I. Mobilization of phosphorus in soil connection with the vital activity of some microbial species. Microbiologiya, 1948; 17 :362–370.
- Ghevariya, K.K., Desai, P.B. Rhizobacteria of sugarcane: In vitro screening for their plant Growth Promoting potentials. Res J Recent Sci., 2014; 3: 52-58.
- Schwyn, B., Neilands, J.B. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem., 1987; 160: 47 56.
- Dobbelaere, S., Croonenborghs, A., Thys, A., Broek, A.V., Vanderleyden, J. Phytostimulatory effect of Azospirillum brasilense wild type and mutant strains altered in IAA production on wheat. Plant Soil, 1999; 212: 155–164.
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., Struhl, K. Current protocols in molecular biology. 1995; Wiley New York.
- Dereeper A., Audic S., Claverie J.M., Blanc G. BLAST-EXPLORER helps you building datasets for phylogenetic analysis.BMC Evol. Biol., 2010; 10: 8-14.
- Hossain, M., Hossain, N., Sultana, F., Islam, M.N., Islam, M., Bhuiyan, K.A. Integrated management of Fusarium wilt of chickpea (Cicer arietinum L.) caused by Fusarium oxysporum f. sp. ciceris with microbial antagonist, botanical extract and fungicide. Afr. J. Biotechnol., 2013; 12(29): 4699-4706.
- Litwin, C.M., Calderwood, S.B. Role of iron in the regulation of virulence genes. Clin. Microbiol. 1993; 6: 137-149.
- Karimi, K., Amini, J., Harighi, B., Bahramnejad, B. Evaluation of biocontrol potential of Pseudomonas and Bacillus spp. against Fusarium wilt of chickpea. Aust. J. Crop Sci., 2012; 6(4): 695-703.
- Moradi, H., Bahramnejad, B., Amini, J., Siosemardeh, A., Haji-Allahverdipoor, K. Suppression of chickpea (Cicer arietinum L.) Fusarium wilt by Bacillus subtillis and Trichoderma harzianum. Plant omic J., 2012; 5(2): 68-74.
- Subhani, M.N., Sahi, S.T., Ali, L., Hussain, S., Iqbal, J., Hussain, N. Management of Chickpea wilt caused by Fusarium oxysporium f. sp. ciceris through antagonistic microorganisms. Can. J. Plant Protec., 2013; 1(1): 1-6.
- Mane, S.S., Pal, M. Screening of antagonists and effects of their cultural filtrate on growth and biomass production of Fusarium oxysporum f. sp. ciceris. J. Plant Dis. Sci., 2008; 3(1): 74-76.
- Arfaoui, A., El Hadram, A., Mabrouk, Y., Sifi, B., Boudabous, A., El Hadrami, I., Daayf, F., Cherif, M. Treatment of chickpea with Rhizobium isolates enhances the expression of phenylpropanoid defense related genes in response to infection by Fusarium oxysporum f. sp. ciceri. Euphytica, 2007; 45: 45-48.
© The Author(s) 2017. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.