ISSN: 0973-7510
E-ISSN: 2581-690X
Plants with medical properties are often enriched with endophytes that have the potential to produce important bioactive compounds. Endophytes after entering the plant tissue may either colonize a particular tissue or may spread throughout the host plant without causing damage. The possession of pharmaceutical and biological properties has made the Holostemma ada-kodien Schult as one of the widely used plants of medicinal importance in India. Following the direct cut method three endophytic bacterial strains (UC H1, UC H4 and UC H7) were isolated, identified and characterized from the healthy looking rhizome of H. ada-kodien. Among these isolates, UC H1 and UC H4 were found to have many properties like antibacterial compounds, hydrolytic enzymes and plant growth promoting traits. The isolate UC H4 have ability for Indole-3-Acetic Acid (IAA) production of 513.54 U/ml and very good protease and pectinase activities of 20.65 U/ml and 16.09 U/ml respectively. So far no reports are available on the endophytic microflora of H. ada-kodien.
Endophytic bacteria, 16S rRNA gene sequencing, antibacterial activity, IAA production, RP-HPLC, phosphate solubilisation, ACC deaminase production
There exists a wide and unique range of interaction between microorganisms that lead an endophytic mode of life with their host plants. The endophytic micro-organisms can alter, develop and establish new environment within the host plant and can develop a new niche for themselves so that the host plants can be considered as a composite micro-ecosystems. For the fine terms of host-entophyte relations, endophytes produce various types of bioactive compounds that are metabolites which can lead to new natural forms with a potential role in agricultural, medicinal and industrial fields. In the common endophytic micro-organisms, bacteria are one of the important groups involving fineness of the plants by protecting from biotic and abiotic stresses, prevention of diseases by producing antagonistic compounds, production of hydrolytic enzymes, boosting plant growth via. chelation of iron, solubilization of phosphate, production of phyto-hormones including auxins, 1-aminocyclopropane-1-carboxylate deaminase enzyme production and so on. Holostemma ada-kodien Schult. is one of the most important laticiferous plant used in Indian Ayurvedic medicines. Rhizome of the plant is often the cure for burning sensation, constipation, cough, fever and stomach pain and it is also used as a rejuvenative drug due to the presence of enriched compounds such as terpenoid sugars, lupeol, amyrin, sitosterol, alanine, aspartic acid, glycine, threonine, serine and valine. There is high possibility for finding endophytic microorganisms in such plants that has ample secondary metabolites in its plant tissue. In addition to this endophytic isolates from medicinal plants may share many properties of its host plant. Therefore the present work was meant the endophytic bacterial isolation from the medicinally important plant H. ada-kodien and further characterization of the same.
Endophytic bacterial isolation
Endophytic bacteria occurring in the healthy looking rhizome of H. ada-kodien were subjected to isolation studies. The collected samples were rinsed in steady flowing water for removing soil and dirt followed by surface sterilization (Sun et al. 2008). The samples were dipped in diluted ethanol (70%) for a period of 1 min, which was then subjected to 5 minutes treatment with 1% of sodium hypochlorite. This followed by subsequent 1 minute treatments and again treated with 70% ethanol. Then the samples were washed several times and the last treated wash was spread on the nutrient agar (NA) plates as the control for confirming that the isolates are surely endophytes Sterilized filter paper assisted the drying of samples which was cut into small pieces of about 1 cm3. The surface of each segments were removed (Islam et al. 2010) and then placed on NA plate and were incubated for 48 hrs at 30°C. The morphologically distinct bacterial colonies were isolated, then sub-cultured for further purification and used for the further studies. The lack of bacterial colonies on the control plates confirmed that the isolated bacteria were endophytes and the surface sterilization programme was effective in removing the contaminants.
Identification of the isolates by morphological characterization and 16S rRNA gene sequencing
The macro-morphological evaluation of bacterial colonies and micro-morphological characterization of bacterial cell was done as per standard protocols in bacteriology.
The identification of isolates was further clarified by using the genetic information of 16S rRNA gene. The forward and reverse primers used were 16S-RS-F (5’CAGGCCTAACACATGCAAGTC3’) and 16S-RS-R (5’GGGCGGWGTGTACAAGGC3’) respectively. The amplification studies were carried out using Applied Biosystems PCR-GeneAmp System 9700, The amplified DNA was checked by electrophoresis using agarose which was then subjected to purification and for sequencing. The sequence quality was checked using Sequence Scanner Software v1 then checked by BLAST analysis to construct phylogenetic tree and viewed in software Phylodraw.
Study for anti-bacterial activity
The anti-bacterial activity was determined using the paper disc susceptibility test. Isolated endophytic bacterial strains were grown in 100ml nutrient broth (peptone: 0.5g; yeast extract: 0.5g and NaCl: 0.5g in 100ml distilled water) and incubated for five days in rotary shaker at 120rpm. The culture media were centrifuged at 10,000rpm for 15 minutes. The supernatant of probiotic isolates were monitored for antibacterial activity against human pathogenic bacteria inoculated on nutrient agar. A total of 20μl of cell-free supernatant was applied on 6mm diameter cellulosic disc on to the lawn of 200µl of indicator bacteria swabbed on nutrient agar along with Amoxicillin as the standard antibiotic and control as nutrient broth. Six bacteria such as Vibrio cholera, Salmonella typhi, Serratia marcescens, Escherichia coli, Bacillus subtilis and Arthrobacter nicotianae were used as indicator organisms. They are cultured in 10ml nutrient broth and incubated for 18hrs at 37°C. The diameter of the inhibition clear zone was measured after 48 hrs of incubation at 30°C.
Study for enzyme production
Primary screening
Primary screening was effected with detection of metabolites like enzymes that have been diffused throughout the agar. The bacteria were screened for cellulase, xylanase, pectinase, protease, lipase, and amylase. The bacterial isolates were cultured on different indicator media for each enzyme study. After the inoculation, the culture was kept in incubator at 30°C for 48 hrs.
Enzyme assay
The enzyme activities were carried out by initially growing the isolates in specific medium for 24 hrs at 120 rpm. After that, 5ml pre-inoculum of each bacteria was transferred to 95ml culture medium with similar composition to pre-inoculum and incubated at 120 rpm for 96 hrs. Samples were collected at 24, 48, 72, 96 hrs and centrifuged at 10000 rpm at 4°C for 20 mins. The cell free supernatant served as crude enzyme. In each enzyme assay, specific substrates were used such as such as casein for protease activity; starch for amylase; CMC for cellulase; pectin for pectinase; p-nitrophenol palmitate (p-NPP) for lipase and xylan for xylanase. Twenty four hour old cultures were transferred to centrifuge tubes and centrifuged at 10000 rpm for 20 min. Cells were discarded and resultant supernatant was used as the crude enzyme for various enzyme assays.
Protease Assay
Protease assay was conducted using casein as the substrate. The tyrosine liberated by the action of enzyme was estimated using Folins reagent at 660 nm. One unit enzyme activity is defined as the amount of enzyme that releases 1μg of tyrosine per ml per minute under the assay conditions.
Amylase Assay
Amylase assay was conducted using starch as the substrate and the reducing sugar liberated was estimated using DNS reagent. The absorbance was measured at 575 nm. Enzyme activity was expressed in units (1 unit/ml = amount of enzyme which releases 1 μ mole glucose under the assay condition.
Cellulase Assay (CMCase assay)
CMCase assay was conducted by using CMC as substrate. Enzyme was added to the substrate and the reaction was proceeded for 10 minutes. Dinitro salicylic acid reagent was used for quantifying the reducing sugar at 540nm.
Pectinase assay
Pectinase assay was conducted using pectin, the substrate pectin was prepared by dissolving 1gm pectin in 100 ml 0.2M phosphate buffer. After the reaction for specified time the end products were measured at 540 nm. Enzyme activity expressed as μmol glucose released per min-1ml-1 of culture filtrate as enzyme solution.
Lipase Assay
Lipase assay was conducted using p-NPP. The absorbance of the products was measured at 410nm. One unit of enzyme activity is defined as the amount of enzyme that released 1μmol p-nitrophenol per minute from the substrate. p-nitrophenol was used as the standard.
Xylanase Assay
Xylanase assay was conducted using oat spelts xylan. The concentration of reducing sugars released was estimated against xylose standard by noting the absorbance at 540nm. 1 unit of xylanase activity was defined as moles of xylose liberated per minute per ml of enzyme preparation.
Study for the screening of plant growth augmenting traits
Screening for Indole 3-acetic acid
Nutrient broth (20 ml) containing L-tryptophan was used for screening bacterial isolates. The bacteria were allowed to grow for period of 10 days and inoculated cultures were subjected to centrifugation at 3000 rpm for 20 mins (Rahman et al. 2010). Supernatant was collected and used for the further analysis. A mixture of 1 ml culture supernatant and 2 ml of Salkowski solution was incubated for 20 minutes in the absence of light. After incubation, the presence of pale red-dark red colour indicated IAA production. The un-inoculated medium served as the control. The mixture was read in spectrophotometer at absorbance of 530 nm. Indole acetic acid was estimated using pure IAA as standard and the values were denoted as mg/ml. In order to confirm the production of IAA positively answered microbes were again subjected to growth in NB supplemented with L-tryptophan. The pH value of this NB preparation was then adjusted to 2.5 using dilute HCl (1N) which was then extracted with ethyl acetate. The extract was vacuum dried and the powder mixed with one ml methanol and subjected to RP-HPLC. The elution was monitored at 280 nm.
Screening for phosphate solubilization
The inorganic phosphate solubilization studies were conducted using the method developed by Jasim et al. (2013). The bacterial cultures were streaked onto modified Pikovaskaya’s nutrient medium consisting of inorganic phosphate, which is then kept at 30°C for 96-168 hours. A yellow halo like zone of clearance around the bacterial colony indicated mobilisation of mineral phosphate. The solubilization index for phosphate mobilization was calculated by
Solubilization index (SI) = (Diameter of Colony + Diameter of halo zone) / Diameter of Colony
Screening for ACC deaminase production
Method of Jasim et al. (2013) was followed for screening the production of ACC deaminase. The microbial cultures were streaked onto Dworkin and Foster salt medium supplemented with 0.2% w/v ammonium sulphate and the petriplates were incubated for a period of 2-3 days at 30°C. The growth bacterial colony indicated production of ACC deaminase.
Newly isolated endophytic bacteria from the healthy looking rhizome of H. ada-kodien were designated as UC H1, UC H4 and UC H7. There was no growth of microorganisms on the control plate. So, it clearly confirmed that all the isolates are endophytes and also all the epiphytic microorganisms were removed at the surface sterilization steps. The endophytes obtained from the isolation were considered for further analyses.
The macro and micro morphological characterization was used for the identification of isolates (Table-1, 2). The isolates UC H1 and H7 and showed positive results for Indole Production test, Starch hydrolysis, Catalase, Voges-Proskauer’s test, Methyl Red test, Nitrate Reduction and Citrate utilisation and negative for Urease test. UC H4 answered positive for Catalase and negative for oxidase test, TSI test (Tripple sugar Iron agar) and Simmons citrate agar test.
Table (1):
Colony characteristics of endophytes isolated from H. ada-kodien.
Sl No. |
Code |
Colony Shape |
Colony Margin |
Colony Elevation |
Optical Density |
Colour |
|---|---|---|---|---|---|---|
1 |
UC H1 |
Round |
Circular |
Pulvinate |
Clear |
White |
2 |
UC H4 |
Round |
Circular |
Pulvinate |
Opaque |
Yellow |
3 |
UC H7 |
Punctiform |
Circular |
Umbonate |
Clear |
White |
Table (2):
Micromorphological characteristics of endophytes from H. ada-kodien.
Sl No. |
Code |
Gram staining |
Cell Shape |
Motility |
Endospore |
|---|---|---|---|---|---|
1 |
UC H1 |
Positive |
Rod |
Positive |
Present |
2 |
UC H4 |
Positive |
Spherical |
Negative |
Absent |
3 |
UC H7 |
Positive |
Rod |
Positive |
Present |
Molecular characterizations of three isolates
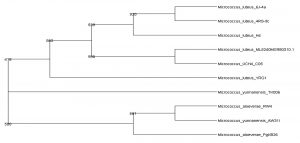
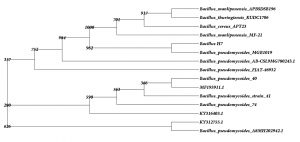
The identification and characterisation of selected isolates were done at the molecular level by amplifying the 16S rRNA gene from each isolate using suitable primers which was then subjected to partial sequencing. The gene sequence of UC H1, UC H4 and UC H7 were 1249 bp, 947 bp and 1243 bp long respectively. The DNA sequence analysis of 16S rRNA gene sequences of UC H1, UC H4 and UC H7 in BLAST showed its maximum similarity of 100% to Bacillus pumilus, Micrococcus luteus and Bacillus pseudomycoides respectively. The tree output was viewed in the tree viewing software Phylodraw (Fig. 1, 2, 3). These bacterial isolates were considered as the endophytes from the rhizome of H. ada-kodien.
Bacillus pumilus INR7, previously isolated as an endophyte from heavily infested cucumber plant has been commercialized as a biological control product against both soil borne pathogens and foliar pathogens. They act by the means of direct antagonism along with the triggering of systemic resistance (Jeong et al. 2014). Symbiotic influence of endophytic B. pumilus in Ocimum sanctum was noted by Murugappan et al. (2013). The survival of B. pumilus as an endophyte in alcohol treated in vitro cultured grapes has been observed (Thomas 2004). The endophytic bacterium Micrococcus luteus has been isolated from the leaves of Phaseolus vulgaris by Costa et al. (2012). M. luteus strain K39 occurring as endophytes in the roots of Cyperus conglomeratus was obtained from the Red Sea shore of the Kingdom of Saudi Arabia. This strain is of common occurrence as an endophytic flora of rice plant (Mano and Moriskai 2008). Micrococcus luteus (NR_075062) was recently isolated from the leaf and stem of Panax ginseng by Hong et al. (2018). B. pseudomycoides was isolated from yam plants (Souza et al. 2017).
Antibacterial activity
Preliminary antibacterial activity of three endophytic bacteria estimated based on the clear zone production reveals that supernatants obtained from all the isolates against the tested bacteria was found to be negative except in the case of Serratia marcescens and Arthrobacter nicotianae. The presence of antibacterial activity of probiotic bacteria is shown in Table-3. The presence of zone of inhibition clearly revealed that Micrococcus luteus and Bacillus pseudomycoides exhibited maximum inhibition against gram positive bacteria A. nicotianae (12mm).
Table (3):
Antibacterial activity of three endophytic bacterial isolates.
| Sl No. | Code | Zone of inhibition in mm | |||||
|---|---|---|---|---|---|---|---|
| Test bacterial strains | |||||||
| Vibrio | Salmonella | Serratia | Bacillus | E.coli | Arthrobacer | ||
| 1 | Control (Amoxillin) | 20 | 31 | 31 | 31 | 31 | 31 |
| 2 | NA | 0 | 0 | 0 | 0 | 0 | 0 |
| 3 | UC H1 | 0 | 0 | 7 | 0 | 0 | 10 |
| 4 | UC H4 | 0 | 0 | 11 | 0 | 0 | 12 |
| 5 | UC H7 | 0 | 0 | 11 | 0 | 0 | 12 |
Information on the antibacterial activity of these two strains against the tested organisms is not reported. So the far related works are used for the specification. Sadrati et al. (2013) reported that all of the selected fungal isolates have potential to produce antimicrobial compounds against Serratia marcescens with inhibition zone (IZ) value in each isolates ranging from 10.7 mm to 14.3 mm. Another report regarding the isolation of endophytic fungi with antibacterial activity from Calotropis procera showed maximum IZ (17.3 mm) with respect to Aspergillus nomius, A. oryzae, Fusarium solani and Curvularia hawaiiensis against S. marcescens (Rani et al. 2017). Reports on endophytic bacteria showing negative results in the antibacterial compound production are many. The bacterial endophyte, B. pumilus and M. luteus isolated from the leaf of Plectranthus tenuiflorus have no potential for producing antibacterial compounds against Salmonellae typhi, Staphylococcus aureus, E. coli, Klebsiella pneumonia and Streptococcus agalactiae (El-Deeb et al. 2013).
Enzyme production
Primary screening
The newly isolated endophytic bacteria were screened for the ability to produce of enzymes like, protease, cellulase, xylanase, pectinase, lipase and amylase, (Table-4). In protease screening, only B. pumilus and M. luteus showed the clear zone of paracasein formation around the spotted isolates. All the three endophytic bacterial strains showed positive result in the screening test for the synthesis of cellulase, xylanase and pectinase and negative result for amylase and lipase. Genera like Bacillus, Paenibacillus and Pseudomonas are highly recognized as the members of protease producers. The endophytic bacteria B. pumilus from Plectranthus tenuiflorus have potential to produce protease, pectinase, xylanase, cellulase and M. luteus from the same plant could produce protease, xylanase and cellulase (El-Deeb et al. 2013). Previous studies suggested that Bacillus sps. are the most important genera for the production of xylanase (Irfan et al. 2016).
Table (4):
Primary screening of three endophytic bacterial isolates.
Sl no. |
Enzyme |
UC H1 |
UC H4 |
UC H7 |
|---|---|---|---|---|
1 |
Protease |
+ |
+ |
– |
2 |
Amylase |
– |
– |
– |
3 |
Cellulase |
+ |
+ |
+ |
4 |
Pectinase |
+ |
+ |
+ |
5 |
Lipase |
– |
– |
– |
6 |
Xylanase |
+ |
+ |
+ |
Enzyme assay
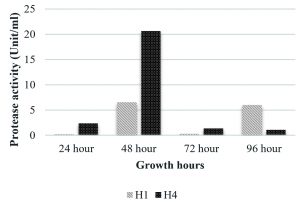
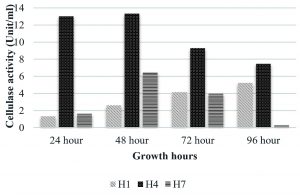
In the present study two strains showed protease production that is B. pumilus and M. luteus (Fig. 4). M. luteus showed the highest activity of 20.65 U/ml after 48 hrs that declined to 1.35 U/ml in the next 24 hrs. B. pumilus also showed the maximum activity of 6.52 U/ml after 48 hrs. Endophytic Bacillus strain from Mangifera indica showed a mild protease activity after 24 hrs i.e. 0.621±0.025; strain from Hibiscus rosa-sinensis had activity that increased with time (0.223±0.014 at 24 hrs; 0.474±0.057 at 48 hrs and 0.576±0.003 at 72 hrs) and an endophytic bacterial strain from Calotropis gigantea with increased activity at 48 hrs that declined at 72 hrs (0.093±0.006 at 24 hrs; 0.577±0.010 at 48 hrs and 0.315±0.013 at 72hrs) was reported by Vijayalakshmi et al. (2016). The growth associated protease production was studied by Chaudhuri et al. (2017) in Bacillus amyloliquefaciens UEF01, an endophyte in the carnivorous plant Utricularia exoleta. Regarding cellulase production (Fig. 5), M. luteus showed the highest activity at 24 hrs and 48 hrs than all other strains i.e. 13.02 and 13.33 U/ml respectively. Ray et al. (2017) observed maximum cellulase yield when the growth of the microorganisms was tested at 40°C, and minimum yield was obtained at 45° C by B. subtilis and B. circulans. The optimum medium conditions for cellulase production by Bacillus subtilis AS3 resulted in comparatively low level of activity with a value of 0.43 U/ml (Deka et al. 2011).
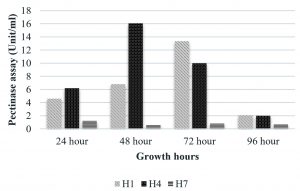
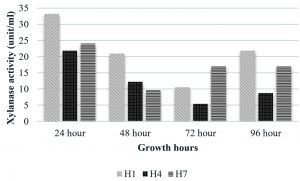
The isolate, UC H7 that was positive in the plate assay for pectinase production did not show considerable activity during the quantitative assay. Pectinase activity of the isolated bacterial strains was comparably negligible. Only B. pumilus and M. luteus showed activity at 72 hrs and 48 hrs respectively (Fig. 6). Paudel et al. (2015) reported that Bacillus sp. HD2 showed maximal PGase activity at 72 hrs. According to Kavuthodi et al. (2015), the required period of incubation time for highest pectinase activity of the isolated B. subtilis was in 72 hrs. These reports are supporting the present work. The enzyme production increased in 60 hrs (1.278 U/ml) and reached the maximum at 72 hrs (1.288 U/ml). El-Deeb et al. (2011) reported the production of pectinase activity by Micrococcus sp., Acinetobacter sp., Planococcus sp., Acetobacter sp. (TUB6) and Methylcoccus sp. (TUB4) isolated from the rose plant. The endophytes in Lonicera japonica such as Bacillus and Paenibacillus, exhibited detectable levels of cellulase and pectinase activities (Zhao et al. 2015). The maximum xylanase activity was observed in B. pumilus at 24 hrs (33.22 U/ml). At 48 hrs the activity became reduced which again got enhanced at 96 hrs (Fig. 7). Archana and Satyanarayana (1997) reported that several species of Bacillus and fungi secrete high amount of extracellular xylanase.
Plant growth promotion traits
Production of Indole 3-acetic acid (IAA)
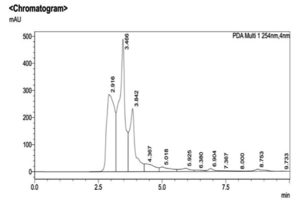
During the primary screening study, IAA production was observed in M. luteus due to the development of red colour in the screening medium on the addition of Salkowski reagent (Fig. 8). The concentration of IAA was calculated by noticing the absorbance at 530nm. It was noticed that maximum IAA production was exhibited by M. luteus (513.54 U/ml). B. subtilis LK14 resulted in considerable level of IAA production with a value of 165.53 µM in 50 ml culture supernatant at the same time B. subtilis LK15 was less efficient with a production value of 32.01 µM in 50 ml culture supernatant (Khan et al. 2016). According to Raddadi (2008) 1.53-9.71 µg/ml IAA was produced by B. thuringiensis. In the present study, confirmation by RP-HPLC analysis ensured the potential of UC H4 to produce IAA into the medium (Fig. 9). El-Deeb et al. 2011 reported that six endophytic bacterial isolates from Rosa damascena trigintipeta had the capacity to produce IAA when the culture medium was supplemented with L-tryptophan, strains TUB3 and TUB5 showed a higher production of IAA (18.6±1.6 and 38.8±3.6 mg IAA/mL, respectively) than TUB1, TUB2, TUB4, and TUB6 (10.2±2.2, 16.6±1.8, 7.8±0.8 mg IAA/mL, respectively). Ribeiro et al. (2018) reported that the bacterial strains isolated from sap, roots and leaves of maize plant effected IAA production with an yield ranging from 3.2 mg/l (Bacillus subtilis) to 61.6 mg/l (Bacillus pumilus ). Similar results were observed in bacteria like Bacillus and Lactobacillus with an IAA production range of 30 to 60 gm/l (Mohite 2013) and 0.81 and 86.82 gm/l (Wahyudi et al. 2011).
Phosphate solubilization
B. pumilus was positive for phosphate solubilization (PS) assay in solid medium (Pikovaskaya’s medium) where in tricalcium phosphate was the phosphate source. Phosphate solubilization capacity was evidenced by the presence of yellow hallow zone of 25mm around the colony after the incubation period (Fig.10). The solubilization index (SI) of B. pumilus was 3.78. Matos et al. 2017 isolated forty endophytic bacteria from banana tree root, out of them Bacillus pumilus EB.102, B. pumilus EB.12, Bacillus sp. EB. 42, Bacillus sp. EB.17, Bacillus sp. EB.81 and B. pumilus EB.05 resulted in the higher levels of solubilized phosphate. B. megaterium and Pseudomonas fluorescens strains showed high phosphorous solubilizing ability (El-komy 2005). The phosphate solubilizing bacteria (PSB) KUPSB12 resulted in phosphate solubilizing halo zone around its colony of 13.0 mm with SI of 2.85 (Paul and Sinha 2016).
Production of ACC deaminase
Among the endophytic bacteria under investigation Bacillus pumilus was positive for ACC deaminase production test in solid medium (Dworkin and Foster salt medium) as shown in Fig.11. Previous reports revealed that various taxa of endophytes especially Bacillus (Xu et al. 2013), Burkholderia (Ali et al. 2014) and Pseudomonas (Qin et al. 2014) have the potential to produce ACC deaminase. Xu et al. (2013) reported that 6% of bacterial isolates from Lycopercium esculentum exhibited an ACC deaminase activity with prominent production by B. subtilis. Thus Bacillus found to be one of the most significant ACC deaminase producing bacteria (Khan et al. 2016).
H. ada-kodien is a very important medicinal plant highly exploited in Indian system of medicine. The rhizome is enriched with terpenoid, lupeol, amyrin, sitosterol, alanine, aspartic acid, glycine, threonine, serine and valine. This is the first report regarding the isolation of endophytic bacteria from Holostemma ada-kodien. Three distinct endophytic bacteria that were isolated from the rhizome of H. ada-kodien Schult, designated as UC H1, UC H4 and UC H7. After the morphological, biochemical and molecular characterization, UC H1 was identified as Bacillus pumilus, UC H4 as Micrococcus luteus and UC H7 as B. pseudomycoides. M. luteus (UC H4) have potential to produce antibacterial compounds, hydrolytic enzymes such as protease, cellulase, pectinase and xylanase as well as IAA. B. pumilus (UC H1) have the ability to produce antibacterial compounds, protease, pectinase, xylanase, ability for phosphate solubilization and ACC deaminase production. B. pseudomycoides (UC H7) have potential to produce antibacterial compounds. From the results it can be concluded that M. luteus (UC H4) and B. pumilus (UC H1) are the most prominent endophytic bacteria with antibacterial activity, enzyme activity and plant growth promoting ability.
ACKNOWLEDGMENTS
The authors are thankful for the facilities provided by the Principal, University College, Thiruvananthapuram, Kerala, India, the Research Centre of University of Kerala. We also express our thanks to Rajeev Gandhi Centre for Biotechnology, Thiruvananthapuram, Nexgene lab, Kochi and Centre for Genomics, Kariavattom Thiruvananthapuram for the DNA sequencing and RP HPLC analysis.
CONFLICT OF INTEREST
Authors declare that there is no conflict of interest
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
- Ali S, Duan J, Charles TC, Glick BR. A bioinformatics approach to the determination of genes involved in endophytic behavior in Burkholderia spp. J Theor Biol. 2014;343:193-198.
Crossref - Archana A, Satyanarayana T. Xylanase production by thermophilic Bacillus licheniformis A99 in solid-state fermentation. Enz Microbial Technol. 1997;21(1):12-17.
Crossref - Chaudhuri M, Biswas S, Pal A, Paul AK. Proteolytic activity of Bacillus amyloliquefaciens UEF01 endophytic to carnivorous plant Utricularia exoleta R. Br. Int J Biotech. 2017;17(3):1-11.
Crossref - Costa LE de O, Queiroz MV de, Borges AC, Moraes CA de, Araujo EF de. Isolation and characterization of endophytic bacteria isolated from the leaves of the common bean (Phaseolus vulgaris). Braz J Microbiol. 2012;43(4):1562-1575.
Crossref - Deka D, Bhargavi P, Sharma A, Goyal D, Jawed M, Goyal A. Enhancement of cellulase activity from a new strain of Bacillus subtilis by medium optimization and analysis with various cellulosic substrates. Enz Res. 2011.
Crossref - El-Deeb B, Fayez K, Gherbawy Y, Elhariry H. Characterization of endophytic bacteria associated with rose plant (Rosa damascena trigintipeta) during flowering stage and their plant growth promoting traits. J Plant Int. 2011;7(3):248-253.
Crossref - El-Deeb B, Fayez K, Gherbawy Y. Isolation and characterization of endophytic bacteria from Plectranthus tenuiflorus medicinal plant in Saudi Arabia desert and their antimicrobial activities. J Plant Int. 2013;8(1):56-64.
Crossref - El-komy HMA. Coimmobilization of Azospirillum lipoferum and Bacillus megaterium for successful phosphorus and nitrogen nutrition of wheat plants. Food Technol Biotechnol. 2005;43(1):19-27.
- Hong CE, Kim JU, Lee JW, Lee SW, Jo I. Diversity of bacterial endophytes in Panax ginseng and their protective effects against pathogens. 3 Biotech. 2018;(8):397.
Crossref - Irfan M, Asghar U, Nadeem M, Nelofer R, Syed Q. Optimization of process parameters for xylanase production by Bacillussp. in submerged fermentation. J Radiat Res Appl Sc. 2016;9(2):139-147.
Crossref - Islam ASM, Math RK, Kim JM, et al. Effect of plant age on endophytic bacterial diversity of Balloon flower (Platycodon grandiflorum) root and their antimicrobial activities. Curr Microbiol. 2010;61:346-356.
Crossref - Jasim B, Jimtha John C, Jyothis M, Radhakrishnan E K. Plant growth promoting potential of endophytic bacteria isolated from Piper nigrum. Plant Growth Regul. 2013;71(1):1-11.
Crossref - Jeong H, Choi S K, Kloepper J W, Ryu C M. Genome sequence of the plant endophyte Bacillus pumilus INR7, triggering induced systemic resistance in field crops. Genome Announc. 2014;2(5):pii: e01093-14.
Crossref - Kavuthodi B, Thomas SK, Sebastian D. Co-production of pectinase and bio-surfactant by the newly isolated strain Bacillus subtilis BKDS1. British Microbiol Res J. 2015;10(2):1-12.
Crossref - Khan AL, Halo BA, Elyassi A, et al. Indole acetic acid and ACC deaminase from endophytic bacteria improves the growth of Solanum lycopersicum. Electronic J Biotechnol. 2016;21:58-64.
Crossref - Mano H, Morisaki H. Endophytic bacteria in the rice plant. Microbes Environ. 2008;23(2):109-117.
Crossref - Matos ADM, Gomes ICP, Nietsche S, et al. Phosphate solubilization by endophytic bacteria isolated from banana trees. Anais da Academia Brasileira de Ciencias. 2017;89(4).
Crossref - Mohite B. Isolation and characterization of indole acetic acid (IAA) producing bacteria from rhizospheric soil and its effect on plant growth. J Soil Sci Plant Nutr. 2013;13:638-649.
Crossref - Murugappan RM, Benazir Beegham SB, Roobia RR. Symbiotic influence of endophytic Bacillus pumilus on growth promotion and probiotic potential of the medicinal plant Ocimum sanctum. Symbiosis. 2013;60(2):91-99.
Crossref - Paul D, Sinha SN. Isolation and characterization of phosphate solubilizing bacterium Pseudomonas aeruginosa KUPSB12 with antibacterial potential from river Ganga, India. Annals of Agrarian Science. 2016;15(1):130-136.
Crossref - Paudel YP, Lin C, Shen Z, Qin W. Characterization of pectin depolymerising exo polygalacturonase by Bacillus sp. HD2 isolated from the gut of Apis mellifera L. Microbiol Disc. 2015;3(1):2.
Crossref - Qin S, Zhang YJ, Yuan B, et al. Isolation of ACC deaminase-producing habitat-adapted symbiotic bacteria associated with halophyte Limonium sinense (Girard) Kuntze and evaluating their plant growth-promoting activity under salt stress. Plant Soil. 2014;374(1-2):753-66.
Crossref - Raddadi N, Cherif A, Boudabous A Daffonchio D. Screening of plant growth promoting traits of Bacillus thuringiensis. Annals Microbiol. 2008;58(1):47-52.
Crossref - Rahman A, Sitepu IR, Tang S-Y, Hashidoko Y. Salkowski’s reagent test as a primary screening index for functionalities of Rhizobacteriaisolated from wild dipterocarp saplings growing naturally on medium-strongly acidic tropical peat soil. Biosci Biotechnol Biochem. 2010;74:2202-2208.
Crossref - Ray AK, Bairagi A, Ghosh SK, Sen K. Optimization of fermentation conditions for cellulase production by Bacillus subtilis CY5 and Bacillus circulans TP3 isolated from fish gut. Acta Ichthyologica et Piscatoria. 2017;37(1):47-53.
Crossref - Rani R, Sharma D, Chaturvedi M, Yadav JP. Antibacterial activity of twenty different endophytic fungi isolated from Calotropis procera and time kill assay. Clin Microbiol. 2017;6(3):280.
Crossref - Ribeiro VP, Marriel IE, Sousa SM de, et al. Endophytic Bacillus strains enhance pearl millet growth and nutrient uptake under low-P. Braz J Microbiol. 2018;49(1):40-46.
Crossref - Sadrati N, Daoud H, Zerroug A, Dahamna S, Bouharati S. Screening of antimicrobial and antioxidant secondary metabolites from endophytic fungi isolated from Wheat (Triticum durum). J Plant Prot Res. 2013;53(2):128-136.
Crossref - Souza JT de, Santos ACOS, Monteiro FP, et al. Genetic diversity and population densities of endophytic Bacillus spp. in yam plants. Bragantia. 2017;76(2):203-208.
Crossref - Sun JQ, Guao LD, Zang W, Ping WX, Chi DF. Diversity and ecological distribution of endophytic fungi associated with medicinal plants. Sci China Ser C. 2008;51:751-759.
Crossref - Thomas P. Isolation of Bacillus pumilus from in vitro grapes as a long-term alcohol-surviving and rhizogenesis inducing covert endophyte. J Appl Microbiol. 2004;97(1):114-123.
Crossref - Vijayalakshmi R, Kairunnisa K, Sivvaswam SN, Dharan SS, Natarajan S. Enzyme production and antimicrobial activity of endophytic bacteria isolated from medicinal plants. Ind J Sci Techn. 2016;9(14).
Crossref - Wahyudi AT, Astuti RP, Widyawati A, Meryandini A, Nawangsih AA. Characterization of Bacillus sp. strains isolated from rhizosphere of soybean plants for their use as potential plant growth for promoting Rhizobacteria. J Microbiol Antimicrob. 2011;3(2):34-40.
- Xu M, Sheng J, Chen L, et al. Bacterial community compositions of tomato (Lycopersicum esculentum Mill.) seeds and plant growth promoting activity of ACC deaminase producing Bacillus subtilis (HYT-12-1) on tomato seedlings. Curr Biol. 2013;30(3):835-845.
Crossref - Zhao L, Xu Y, Lai X-H, Shan C, Deng Z, Ji Y. Screening and characterization of endophytic Bacillus and Paenibacillus strains from medicinal plant Lonicera japonica for use as potential plant growth promoters. Braz J Microbiol. 2015;46(4):977-989.
Crossref
© The Author(s) 2020. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.